BIOL 215 Unit 1
1/109
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
110 Terms
atom
-fundamental unit of elements
-protons + neutrons + electrons
-not all atoms have neutrons --> Hydrogen
atomic number
-determined by # of protons in an atom
important elements making up living organisms
hydrogen, carbon, nitrogen, oxygen make up > 96% of all living organisms
Na, Mg, K, Ca, P, S, and Cl are other important elements
molecule
-made up of 2 or more atoms held by covalent bonds
inner shell
-electrons closest to the positively charged nucleus are attracted mostly strongly to it
-2 electrons max
second shell
-farther away from the nucleus
-can hold up to 8 electrons
atoms with incomplete outer shells (<8 electrons)
- strong tendency to interact with other atoms to either gain or lose enough electrons to fill the outermost shell
covalent bond
- formed when 2 atoms share a pair of electrons
- their negative charges are attracted to the positive charges on both nuclei
electronegativity
-tendency for an atom's nucleus to attract shared electrons in a covalent bond of all living organisms
- higher the electronegativity = stronger the atom attracts electrons
- O > N > C = H
electronegativity trends
left to right: increases bc increased number of protons as the atomic number increases = pull on the electrons
top to bottom: decreases bc of increasing size of the atoms = larger distance = protons less effectively pull on electrons
polar covalent bond
-asymmetry in position of shared electrons bc of differences in electronegativities
nonpolar covalent bond
-equal sharing of electrons because of roughly equal electronegativities
polar molecule
- molecule with an unequal distribution of charge, resulting in the molecule having a partial positive end and a partial negative end (the more electronegative atom)
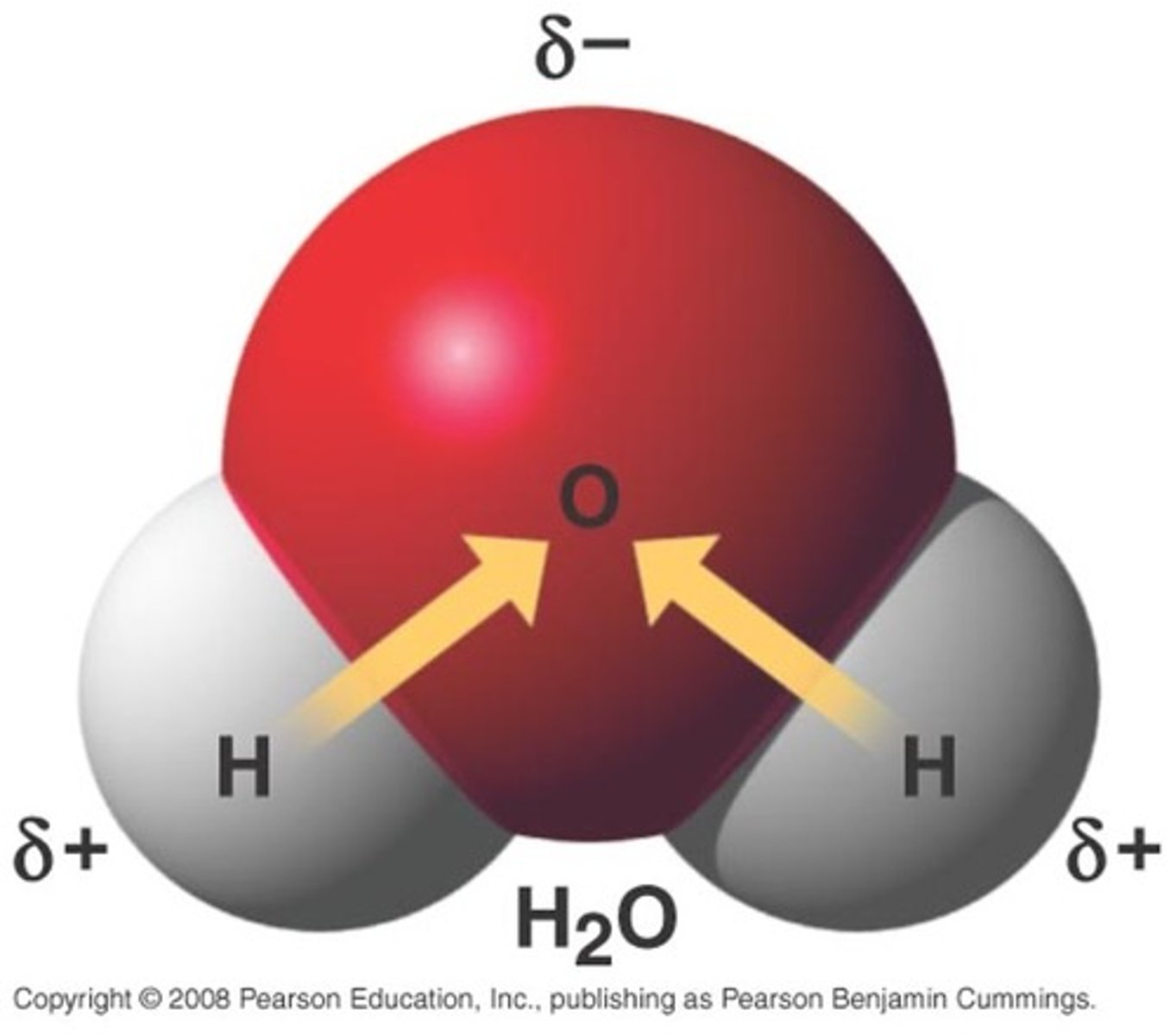
water molecule
- uncharged, but its electrons are unevenly distributed
- the unpaired electron in hydrogen pairs with one of the unpaired electrons in oxygen's outer shell to form a covalent bond
- the electrons in water's covalent bonds are pulled A LOT harder by the oxygen nucleus than they are by the hydrogen nucleus
- oxygen atom pulls bonding electrons so close, acquired a partial negative charge
- electrons pulled away from each hydrogen nucleus = acquire partial positive charge
- water can form hydrogen bonds with oxygen or nitrogen atoms in any molecule = excellent solvent
hydrogen bond
-attraction between partial positive charge on hydrogen atom and partial negative charge on another atom (ex: O, N)
-relatively strong, but not as strong as covalent bonds
-type of intermolecular (between mlcs) interaction
-contribute to secondary/higher order structures of biomolecules
-polar covalent bonds are required for hydrogen bonds to occur
biomolecules
-molecules produced by living organisms that are essential to one or more biological processes
hydrophilic
-mlc that interacts w/ partial charges on water
-likes water
hydrophobic
-mlc that has mostly nonpolar covalent bonds
-can't interact w/ partial charges of water
-doesn't like water
acid
-ion/mlc that releases proton (H+)
base
-ion/mlc that accepts proton (H+)
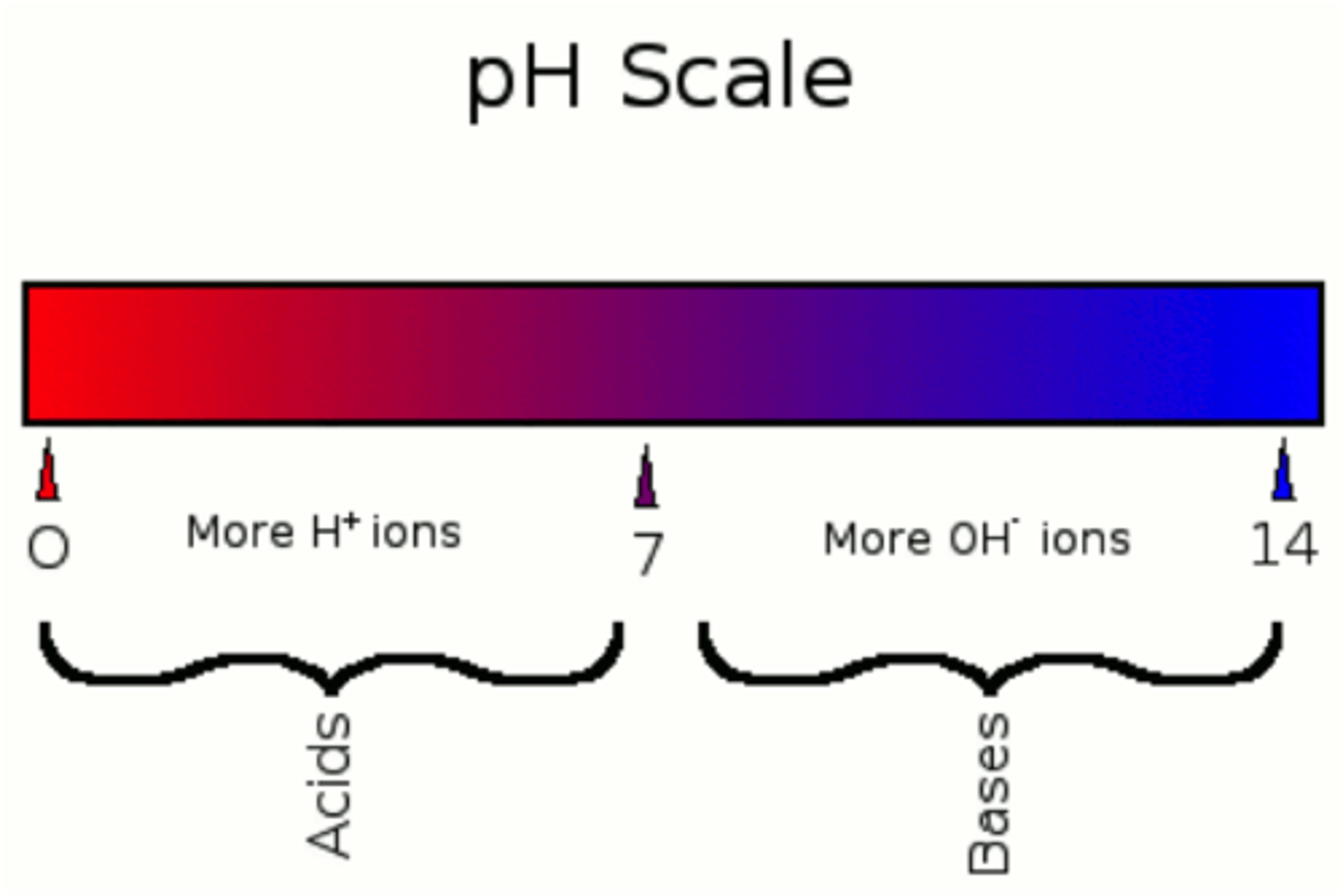
ph scale
-measures concentration of protons in solution
nucleic acid
-made up of a string of nucleotides joined covalently
charge of DNA/RNA
negative
nucleotides
-subunits of nucleic acids
-has phosphate group (in de-protonated form because pKA low), 5-carbon sugar, and nitrogenous base (acts as a base bc picks up proton in water)
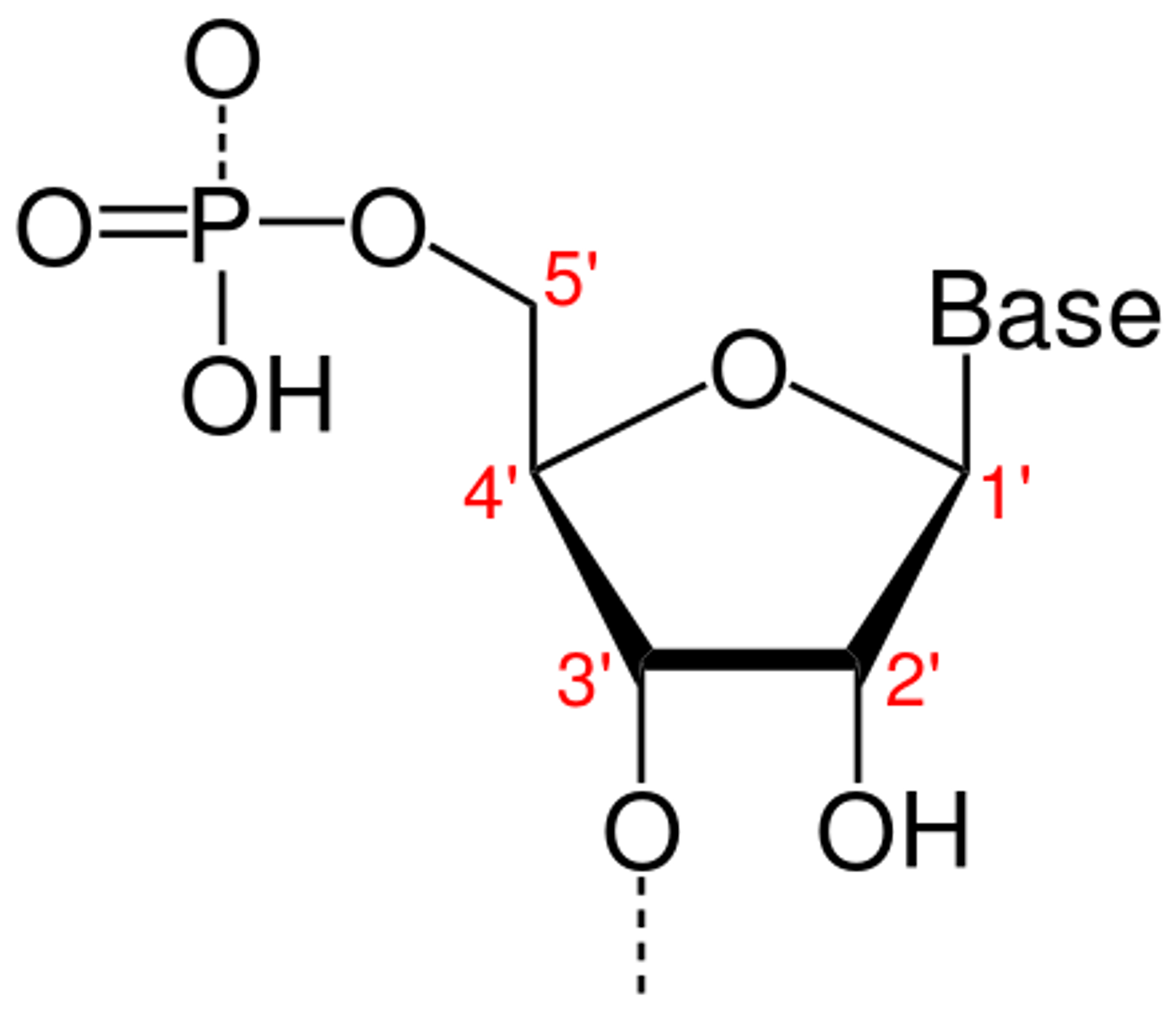
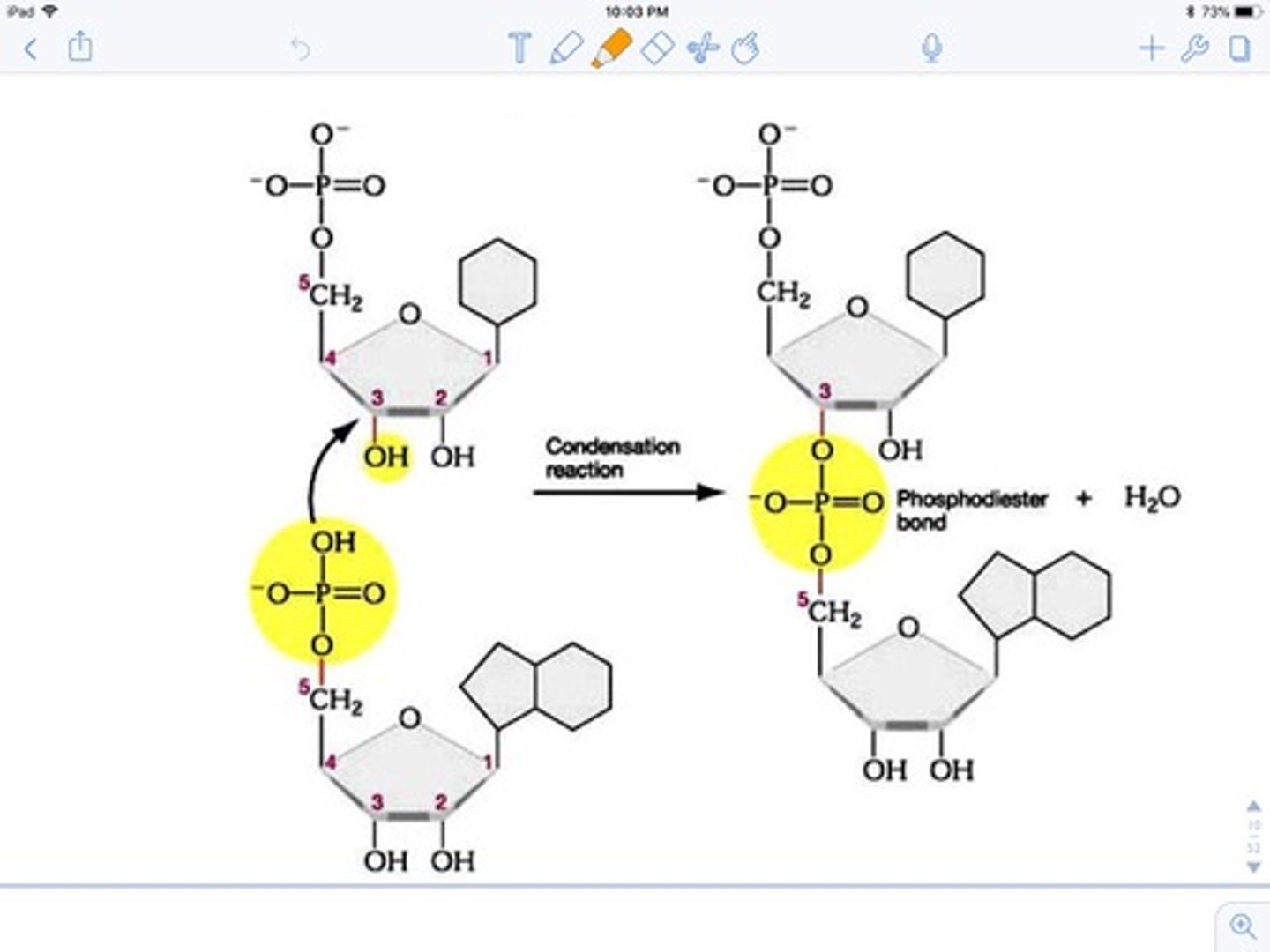
phosphodiester bond
-covalent bond between phosphate group of 5' carbon of one nucleotide and -OH group of 3' carbon of 2nd nucleotide
primary structure of DNA/RNA
-sequence of nucleotides
sugar-phosphate backbone
-"spine" of 5-carbon sugars and phosphate grps in a nucleic acid
-bases project from the backbone
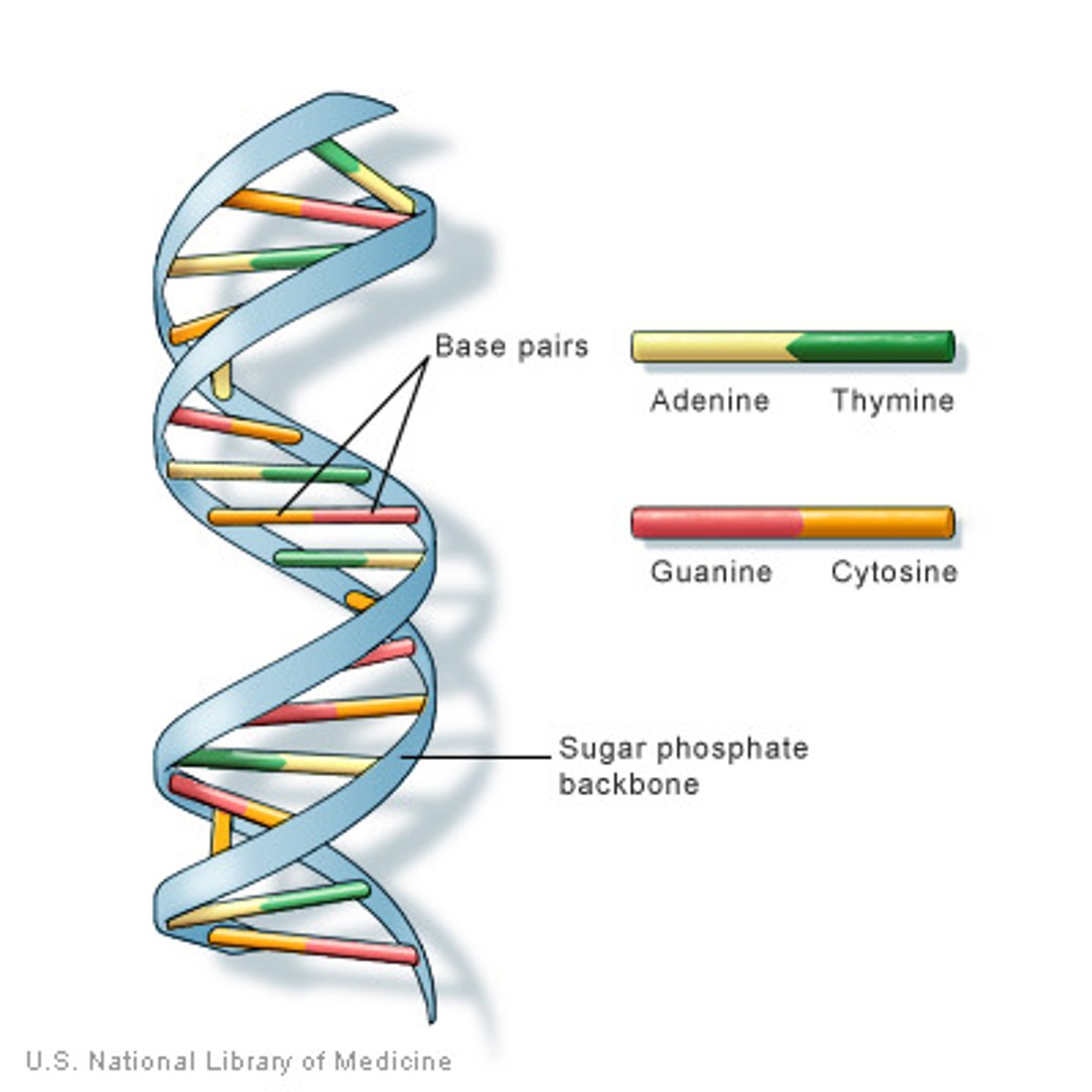
complementary base pairing
-hydrogen bonding between bases
-A +T (2 hydrogen bonds) and C + G (3 hydrogen bonds)
-complementary base pairs are packed in energetically most favorable arrangement in the interior of the double helix (right-handed double helix, one complete turn every 10 base pairs)
-N/O atoms carry partial negative charge while hydrogen atoms they are bonded to carry partial positive charge
secondary structure of DNA/RNA
DNA: formation of helix w/ major and minor groove --> so proteins can get in for replication/transcription
RNA: stem-and-loop structure based on complementary base pairing + antiparallel single strand of RNA, less stable + do more interesting chemistry by of 2' OH group
-sugar phosphate groups are on the outside of the double helix; nitrogenous bases in the inside
why hydrogen bonds for bases instead of covalent bonds?
hydrogen bonds easier to break to open up the strands for replication/transcription
the phosphodiester bonds will hold mlc together even if DNA unzipped
tertiary structure of DNA
-DNA must be tightly packaged to fit in nucleus + but also accessible for gene expression
-DNA folded around 8 histone proteins = nucleosome complexes = control access of proteins to DNA
-chromatin: DNA bound to both histone and non-histone proteins
-these proteins bind to DNA + fold it --> series of organized coils/loops and prevent DNA from tangling
-if DNA needs to be transcribed, nucleosomes slide down DNA to open a region for RNA polymerase to start transcription
-Transient transfection (foreign DNA is introduced into cells and expressed temporarily, without becoming a permanent part of the cell's genome) of cells is known to be more efficient with highly supercoiled DNA compared to linear DNA.
-supercoiled RNA best to reach perinuclear region of cell than linear/relaxed circular DNA = increases transfection efficiency
-linear DNA vulnerable to exonucleases, causing fragments to quickly degrade, where circular DNA is not
topology
geometric/spatial properties of DNA
ex: linear, relaxed, circular, supercoiled
tertiary structure of RNA
-several stem-and-loop sections twist and fold across each other to form a new 3D shape
similarities between DNA/RNA
-primary structure = seq. of nucleotides joined by phosphodiester bonds
-strand have 5' to 3' polarity + sugar-phosphate backbone
-antiparallel structures held together by hydrogen bonds from base pairing
-both have nitrogenous bases + phosphate groups
-both have 3' OH group --> forms phosphodiester bonds
differences between DNA/RNA
-5 carbon sugar in RNA = ribose DNA = deoxyribose
-ribose has OH group bonded to 2' carbon, DNA doesn't
-secondary structure more extensive in DNA than RNA
OH group on 2' of RNA
-more reactive/less stable bc will have partial charge diffs --> the positive H can easily interact w/ other mlcs
pyrmidines
cytosine, thymine, uracil
single ring
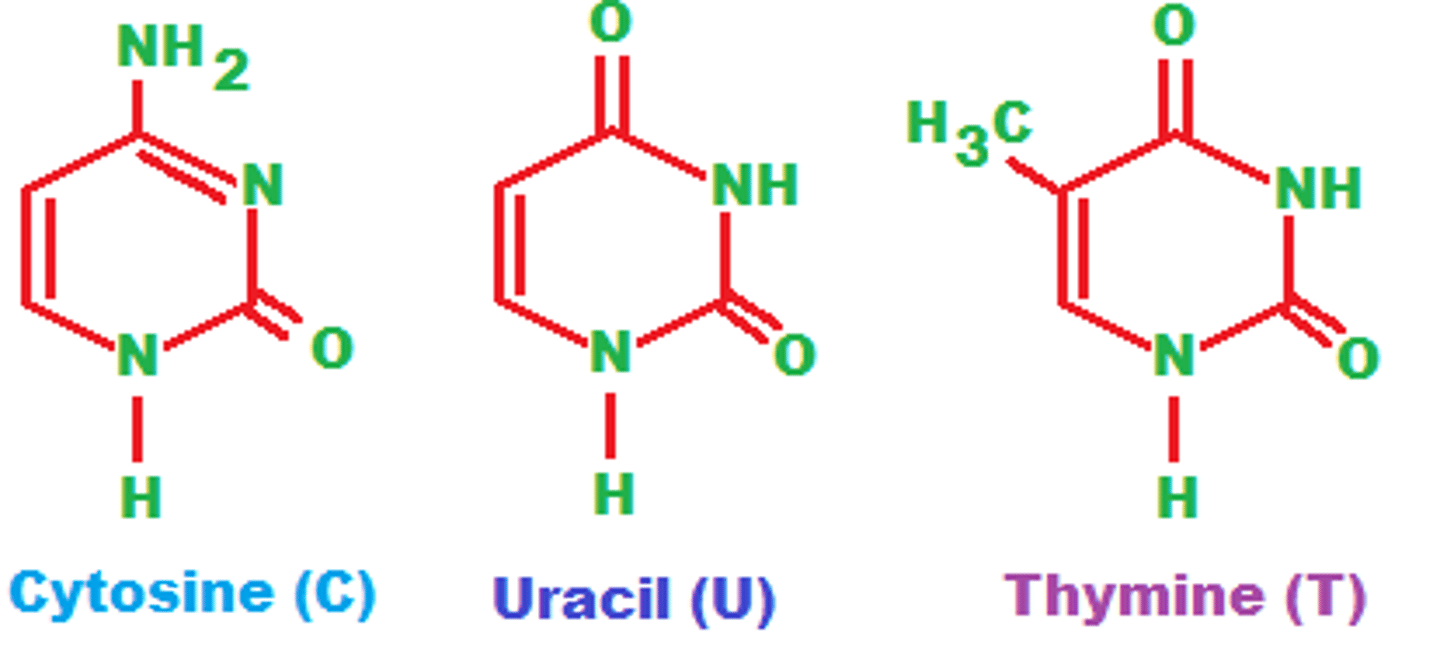
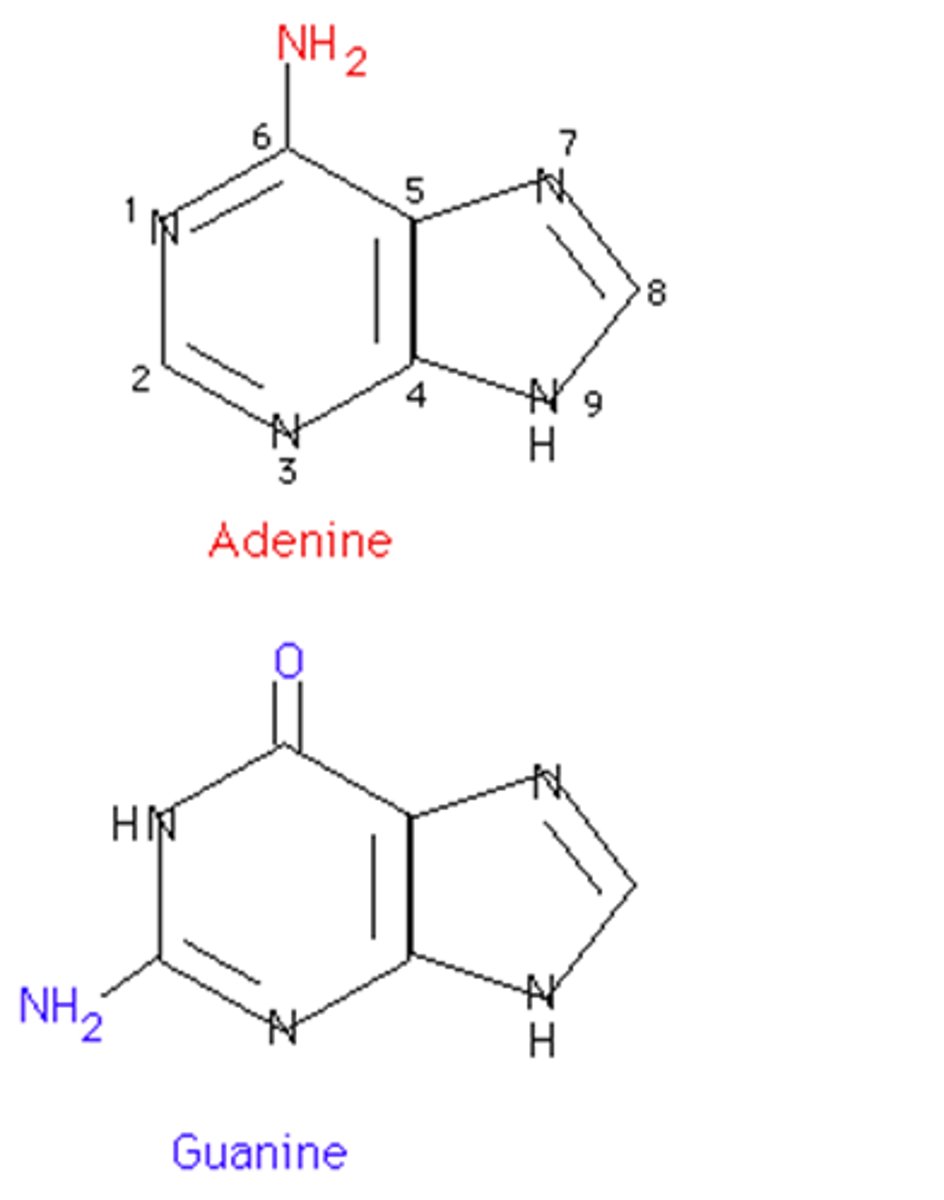
purines
adenine and guanine
double ring
condensation rxn
nucleotides added together bc of OH on 3' and OH on phosphate group --> release water as the 2 nucleotides bond together
state of DNA
permanent, stable
state of mRNA
transient = short-lived bc single strand, less stable bc of 2' OH, more susceptible
the advantage is that once its translated it will be broken down so extra of the protein isn't made
state of proteins
long or short lived, depends
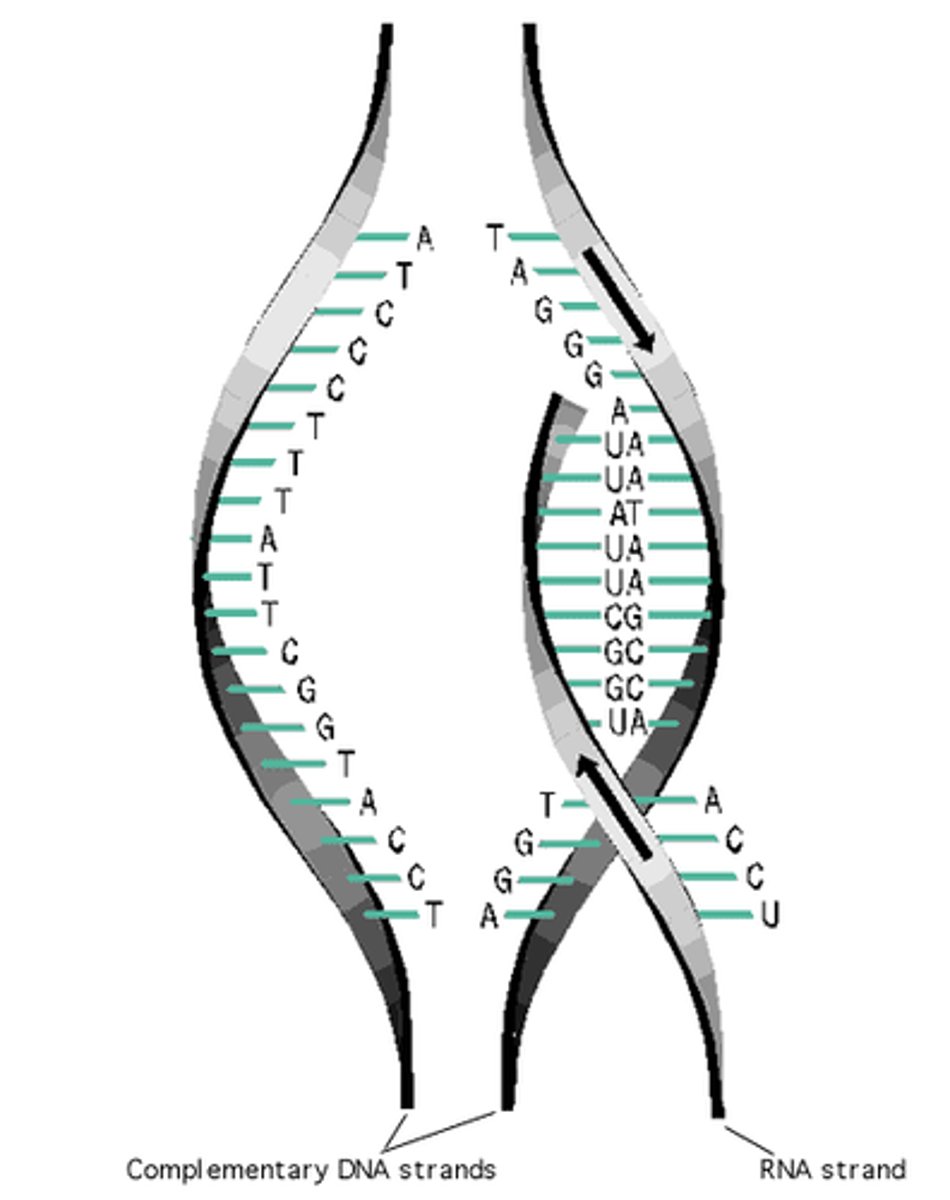
RNA polymerase
- enzyme that catalyzes formation of phosphodiester linkages between ribonucleotides that are complementary to the deoxyribonucleotides in the gene--> forms the mRNA
- recognizes promoter upstream of the gene (TATA)
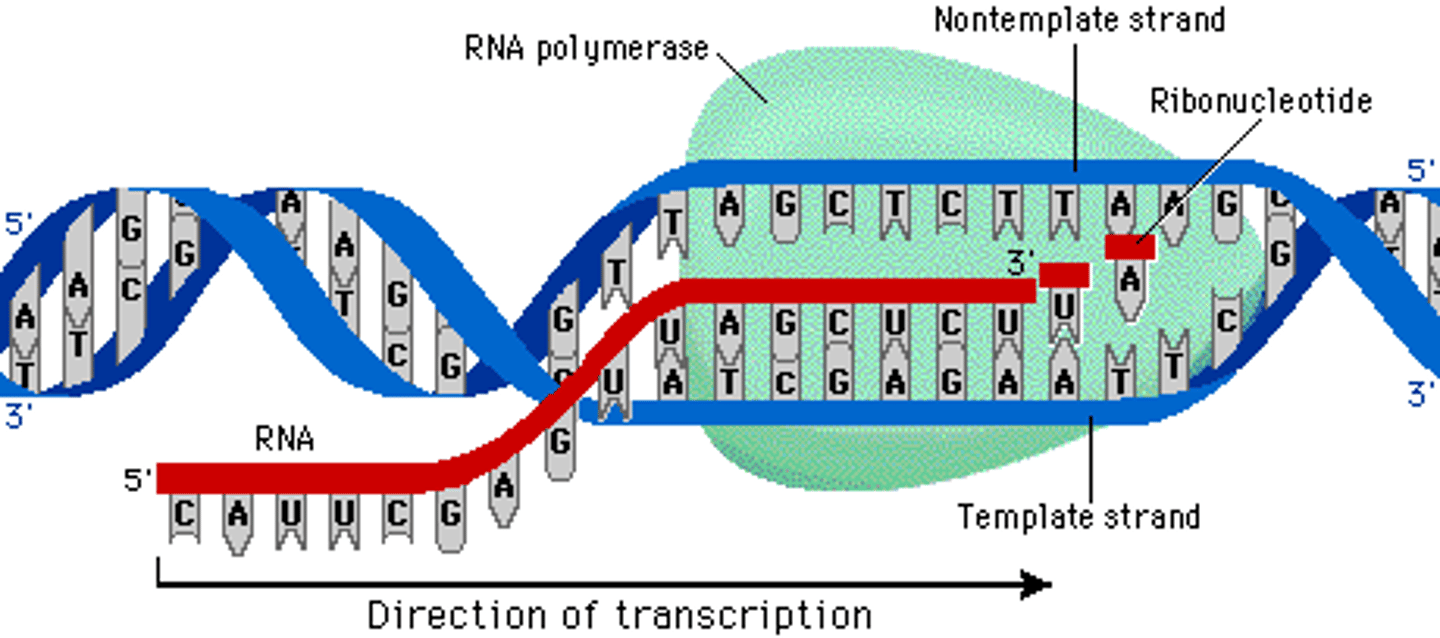
coding strand strand
the strand of DNA that is not used for transcription and is identical in sequence to mRNA, except it contains uracil instead of thymine
both the coding strand + mRNA will be 5' to 3' = in the same direction
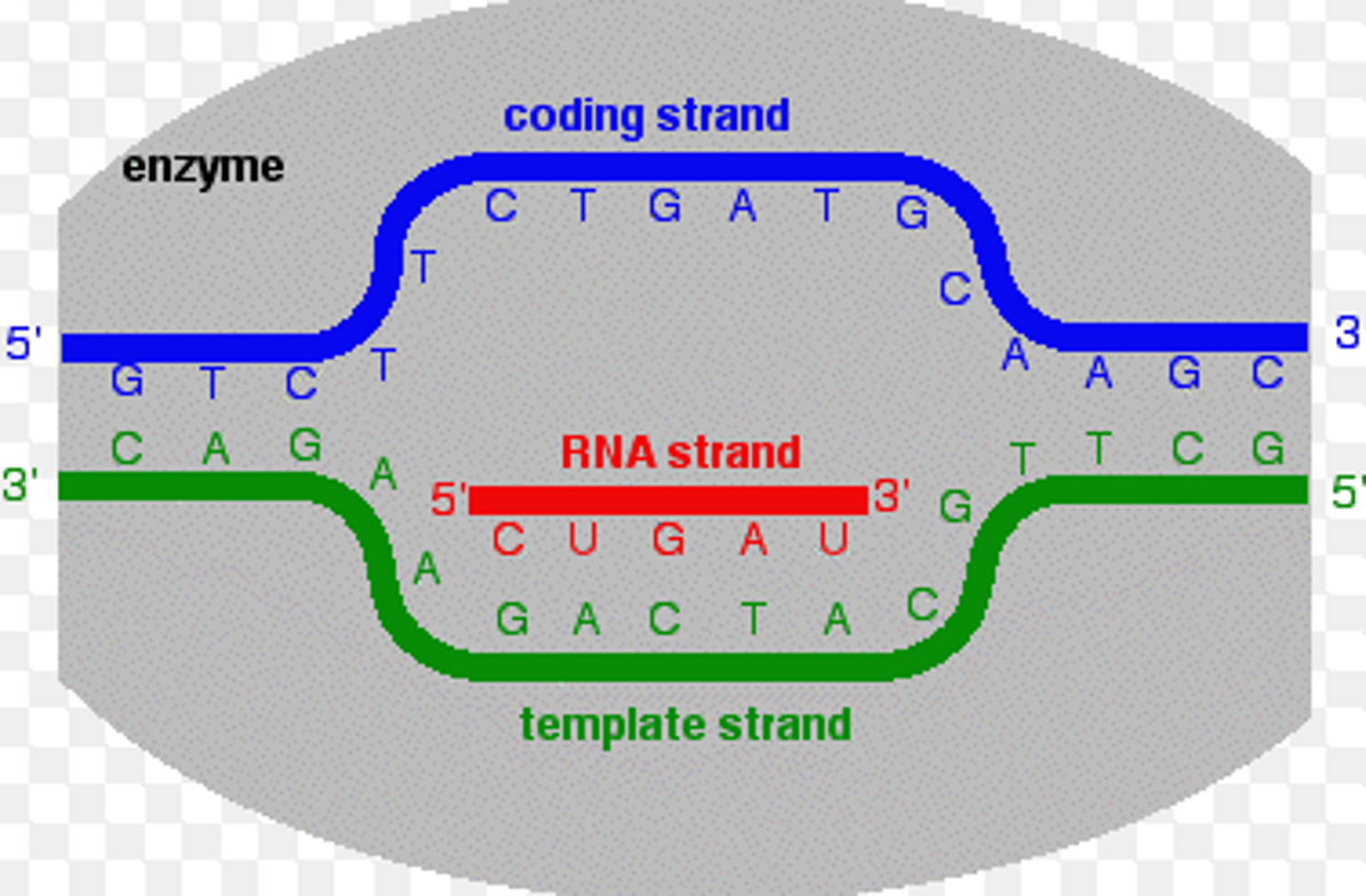
template strand
-read by RNA machinery, converted to mRNA
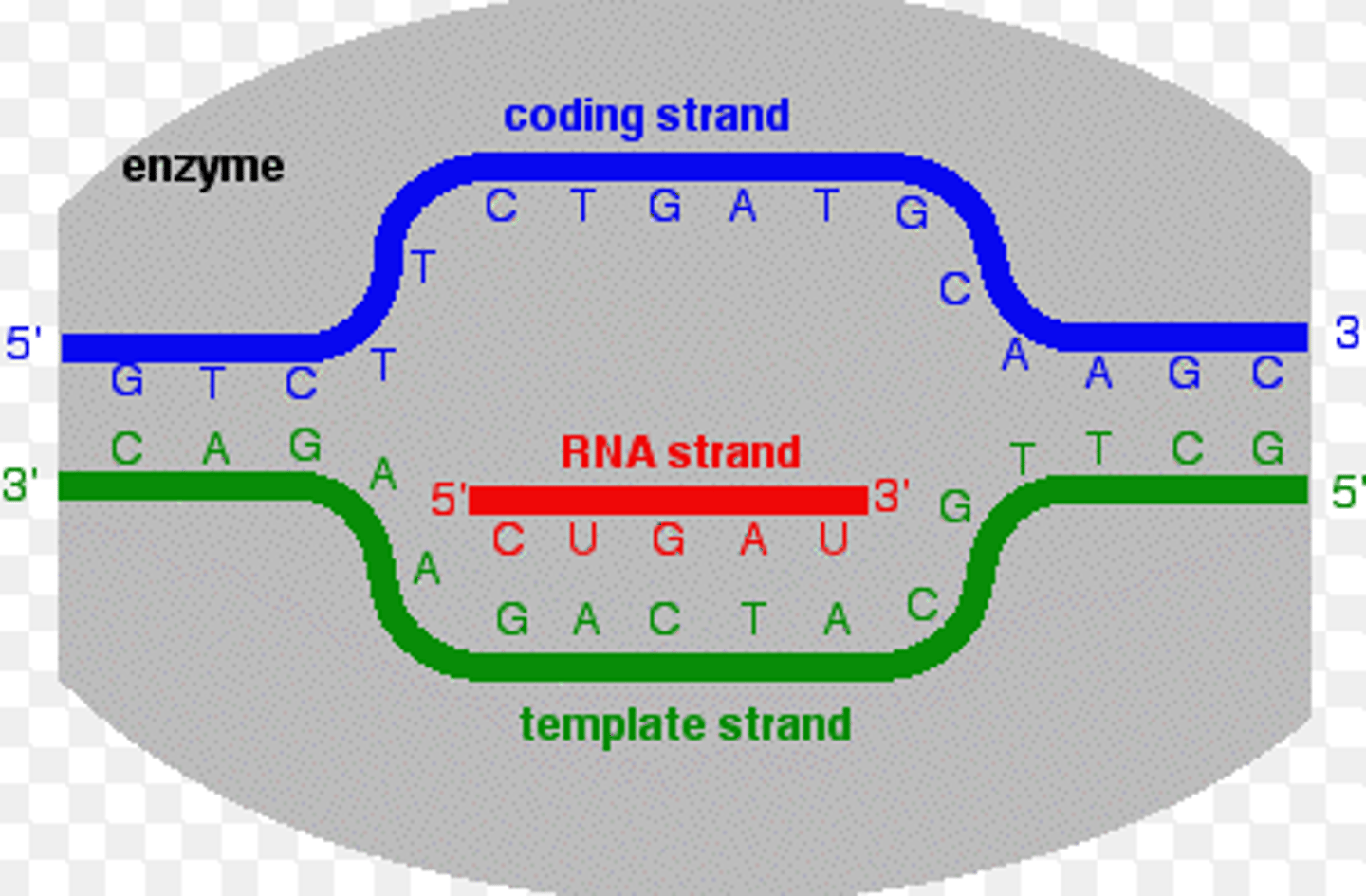
transcription
- happens in nucleus
- signals indicate when mRNAs need to be made
- DNA is the template for the new mRNA
- nucleotides always added to 3' end of growing mRNA chain
- mRNA processed + exits the nucleus for translation into protein
- spontaneous bc substrates are ribonucleotide triphosphates (NTPs) = have a high potential energy
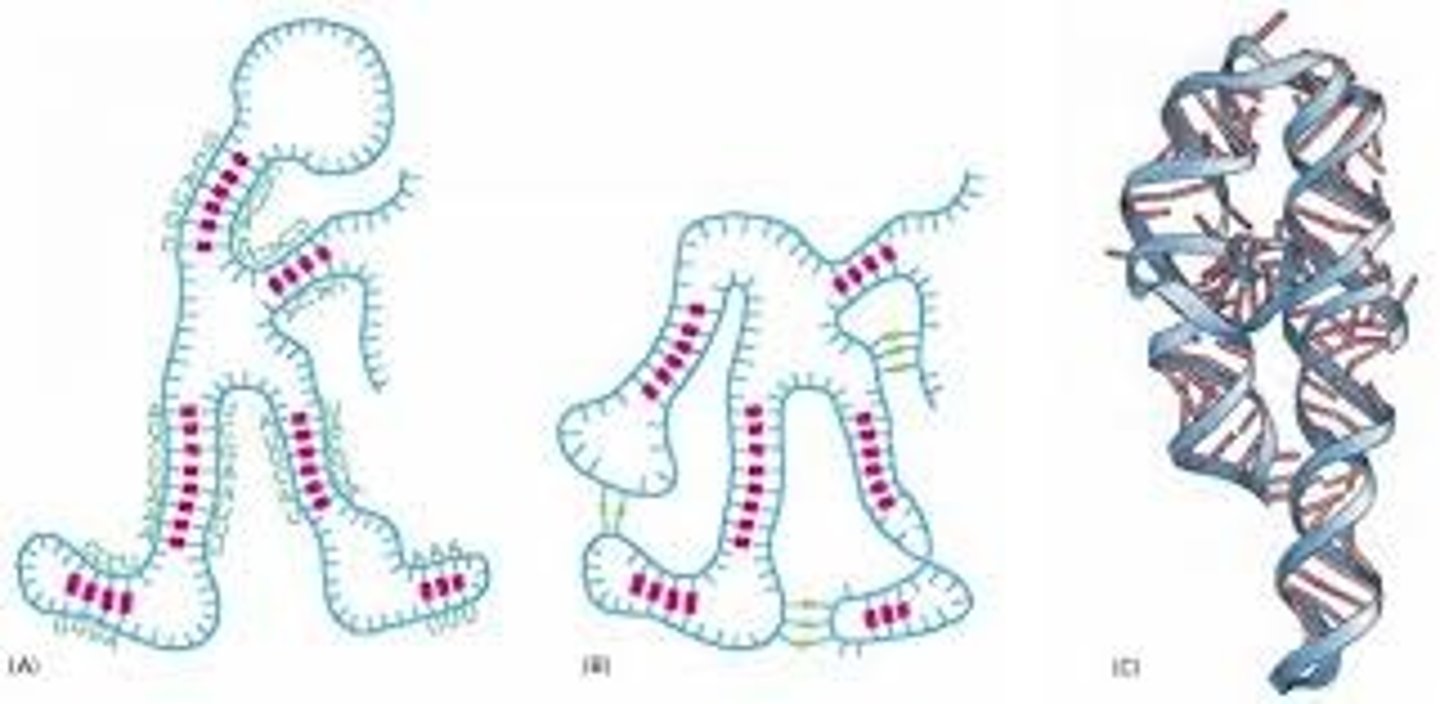
nonconventional base pairing in RNA + unpaired bases
- hydrogen bonding of unconventional base pairing + other intermolecular forces allow RNAs to make a 3D shape
C-U and G-A --> not normal, may just be 1 hydrogen bond
- these forces allow stem and loop structures come together and maintain that association in the tertiary structure = drive geometry
- unpaired bases leave functional groups free to interact w/ other molecules
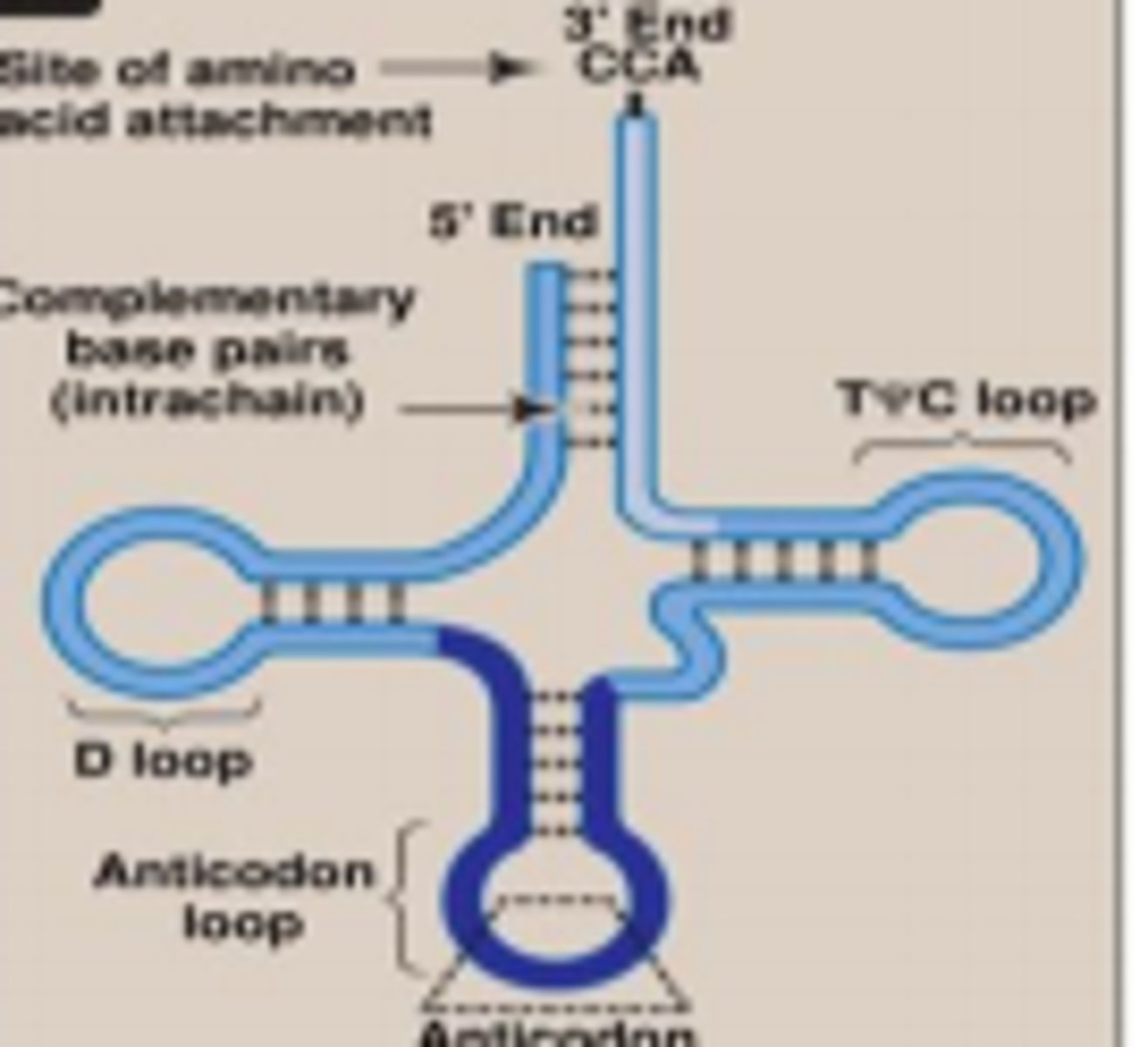
secondary structure of tRNA
- 4 regions of double-stranded RNA --> 3 stem and loops
- T-loop: stabilizing loop structure + proper tRNA folding
- anticodon loop: base pairs w/ codon in mRNA bc has anticodon, creates physical link between mRNA and correct amino acid. hypermodified purine right after the anticodon to prevent this from base-pairing w/ the codon in mRNA = align the codon/anticodon together
- D-loop: region that is recognized by the enzyme aminoacyl-tRNA synthetase
- amino acid attachment on the top of the 3' CCA tail
tertiary structure of tRNA
L shape, formed by 3D folding
aminoacyl-tRNA synthetase
- need to charge appropriate tRNA w/ its amino acid through covalent bond formation
- aminoacyl-tRNA synthetase will recognize the anticodon + allow tRNA to bind
- aminoacyl-tRNA synthetase also has a binding pocket for the side chains of the amino acid
- then uses ATP to form high-energy covalent bond between carboxyl group of amino acid + 3' end of tRNA (CCA tail) that has the free hydroxyl group
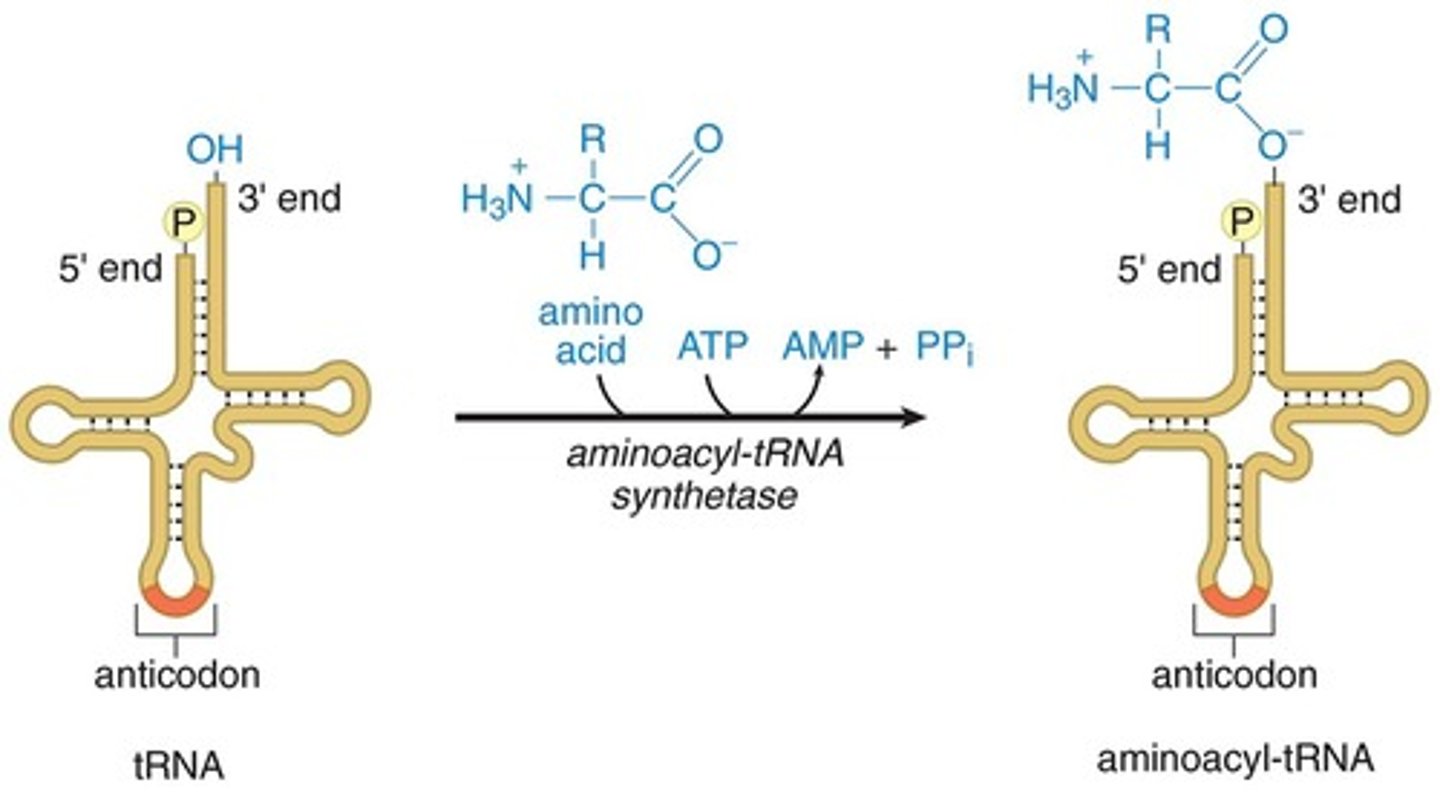
tRNA vs aminoacyl-tRNA
- tRNA = uncharged, no amino acid
- aminoacyl-tRNA = charged, connected to amino acid by high energy bond
how many tRNA synthetases found + why
- 20 enzymes bc 20 amino acids
- recognition of the amino acid side chain is very specific
- but recognition of the anticodon region is less specific/some flexibility
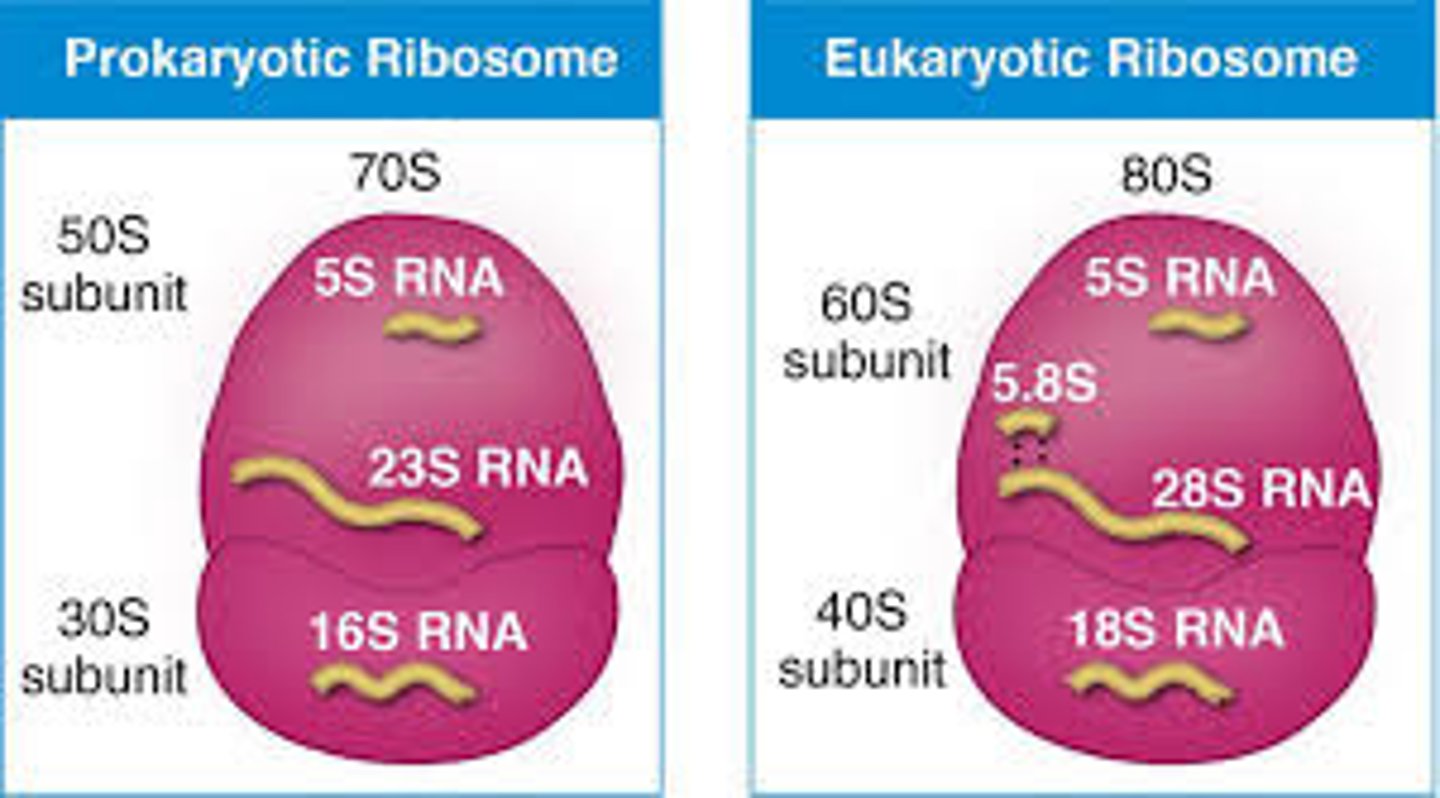
ribosomes
- highly conserved protein-making machines
- composed of rRNA (only in euks, 2/3) and protein (1/3)
- S value: measure of particle size/shape based on its sedimentation rate under acceleration (not additive)
- small subunit: mediates interactions between mRNA and tRNA
- large subunit catalyzes peptide bond formation + has exit tunnel through which growing polypeptide emerges (target for antibiotics)
ribozyme
- made up of rRNA that work together to catalyze peptide bonds
- catalytic activity mediated by rRNAs = speed up chemical rxns
- RNA molecules that act like proteins
- when ribosomes use its rRNA to catalyze peptide bond formation = that rRNA is acting like a ribozyme
ribosomal RNA (rRNA)
RNA molecules that form part of the ribosome
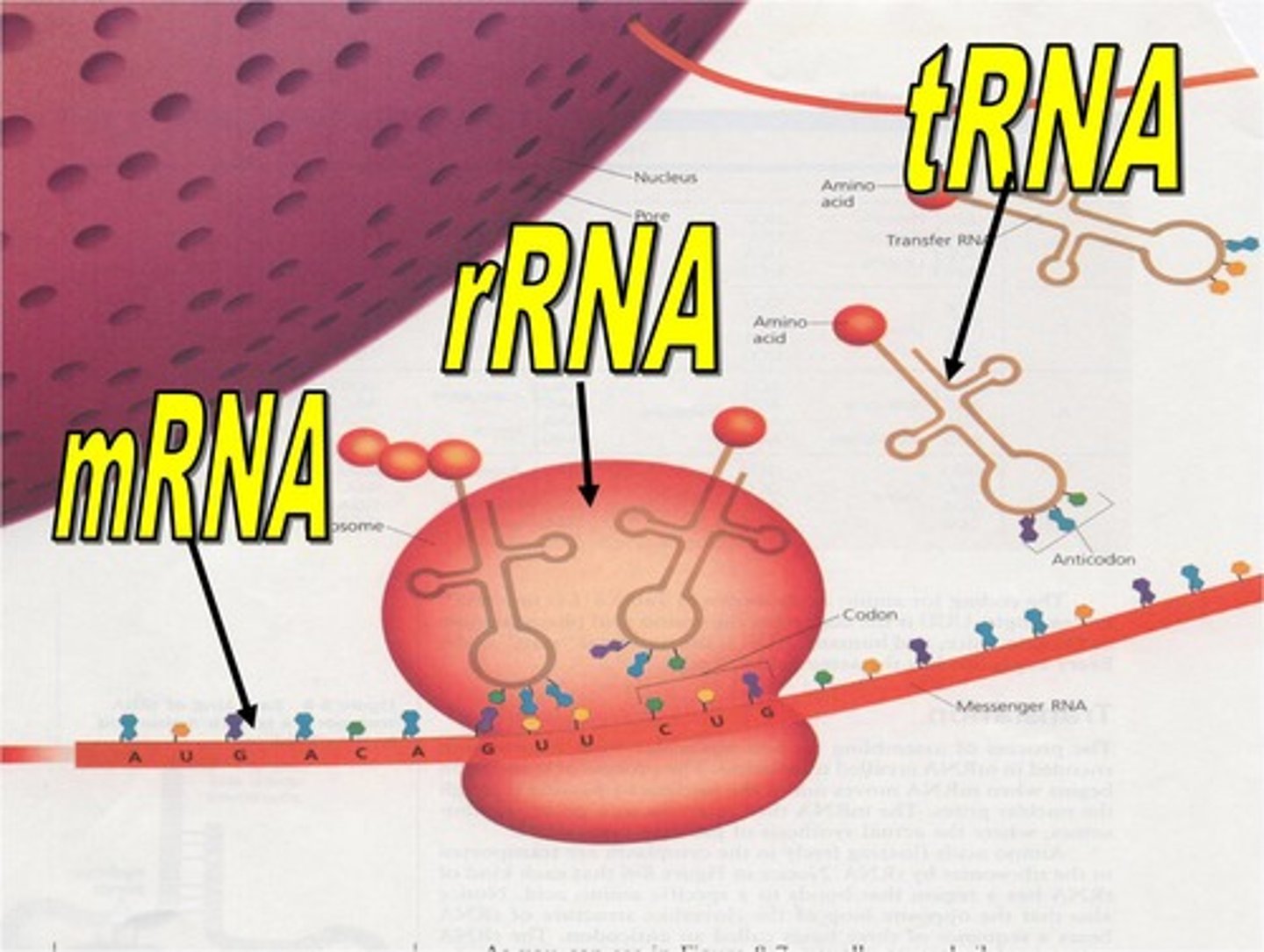
transfer RNA (tRNA)
RNA molecules that function in transferring amino acids to the ribosome during translation
translation initiation
- small ribosomal subunit recognizes the 5' UTR + associates w/ it bc tells the small subunit to bind to the mRNA while bringing in translation initiation factors, initiation tRNA (has Met)
- small subunit will work its way down the mRNA until it reaches AUG = binds, so translation initiation factors dissociate
- large subunit recruited + aligns itself so the Met tRNA is in the P site
- then charged tRNA comes into A site, the Met and new AA will form peptide bond catalyzed by the large subunit
A site
- where amino acid-charged tRNAs enter the ribosome
- if the anticodon of a tRNA binds to the exposed codon in the mRNA = the tRNA and mRNA bind together
P site
- where peptide bond formation takes place
- peptide bond forms between amino acid held by the tRNA in the P site and the amino acid held by the tRNA in the A site
E site
- where tRNAs sit once the amino acid they were carrying has been added to the growing protein
- now uncharged tRNA exits the ribosome from this site
translation elongation
- charged tRNAs enter the acceptor site + the anticodon on the tRNA base pairs w/ the codon in the mRNA
- peptide bond formation between amino acids in the P site
- large subunit translocates (moves down mRNA) towards 3' end by 3 nucleotides, opening up the A site again (where amino acid-charged tRNAs enter the ribosome)
- elongation factors uses GTP (energy) to move ribosome down 3 bases
- ejected now-uncharged tRNA from E site
translation termination
- no tRNAs that recognize the stop codon
- release factors bind in A site once stop codon in A site
- release factors catalyze hydrolysis of last amino acid from its tRNA + requires GTP = this hydrolysis releases free carboxyl terminus (C-term) of the protein
- released polypeptide chain
- ribosomal subunits are then reused, not broken down using energy from GTP hydrolysis
- additional factors use energy in GTP hydrolysis to disassemble the large and small ribosomal subunits + mRNA
can there be many initiation sites on mRNA
yes, many rounds of initiation occur on mRNA = make lots of the protein
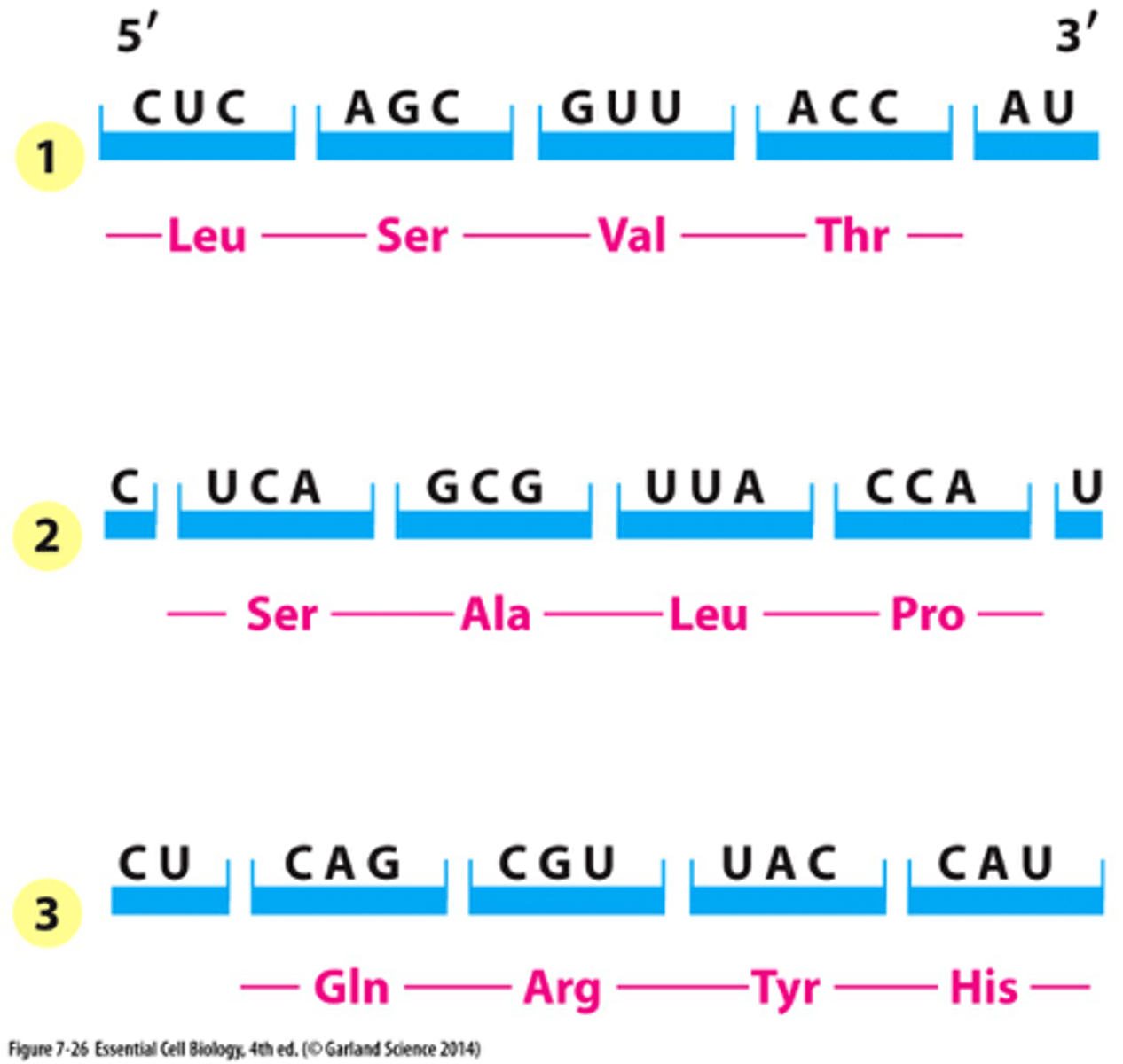
genetic code being unambiguous
- each codon codes for one amino acid
genetic code being redundant
- multiple codons per amino acid
genetic code being conservative
- 3rd codon least important
mRNA reading frames
3 reading frames, only one of these reading frames encodes the actual message because the other ones don't have start codon or encounter stop codons too quickly
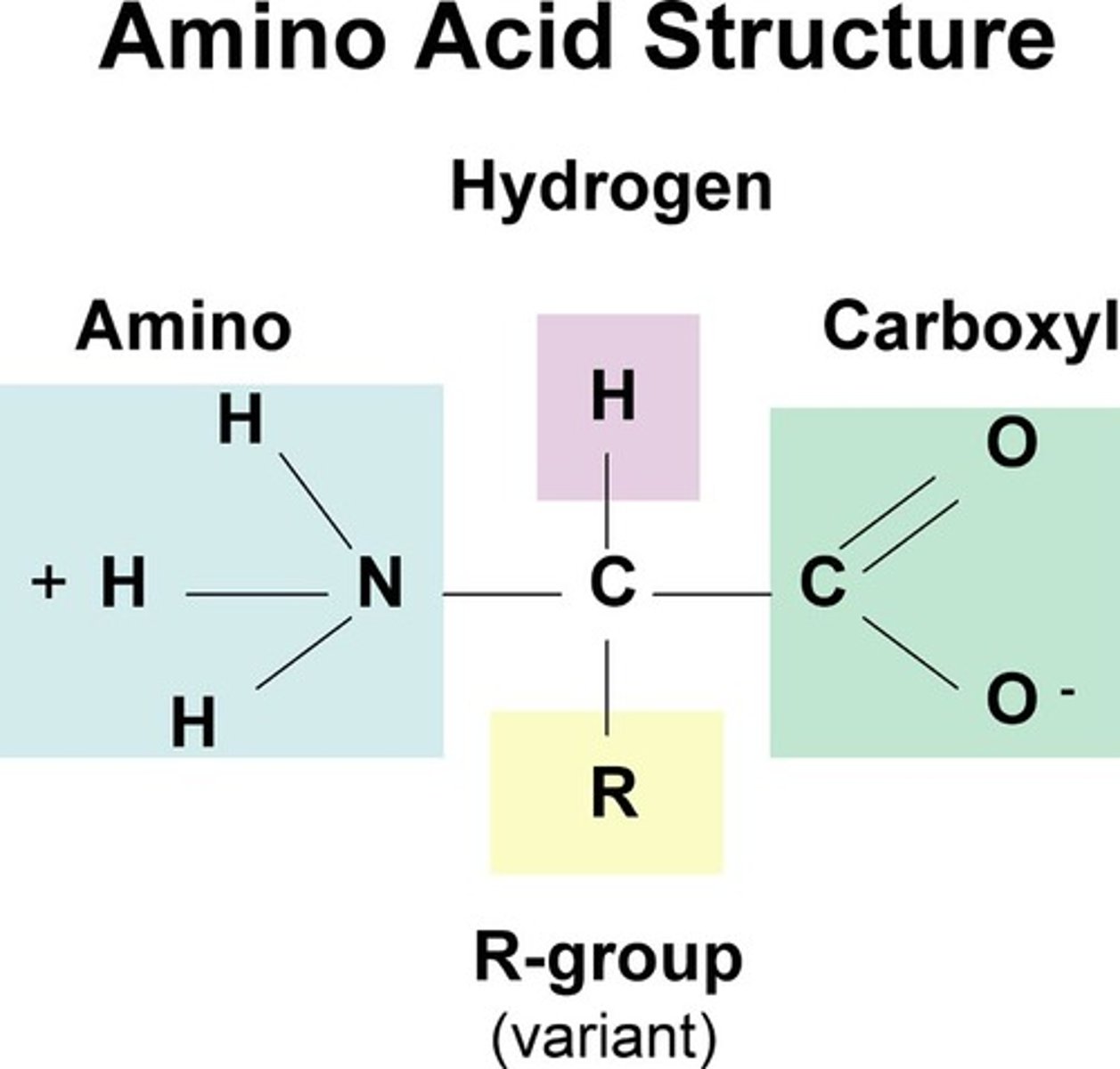
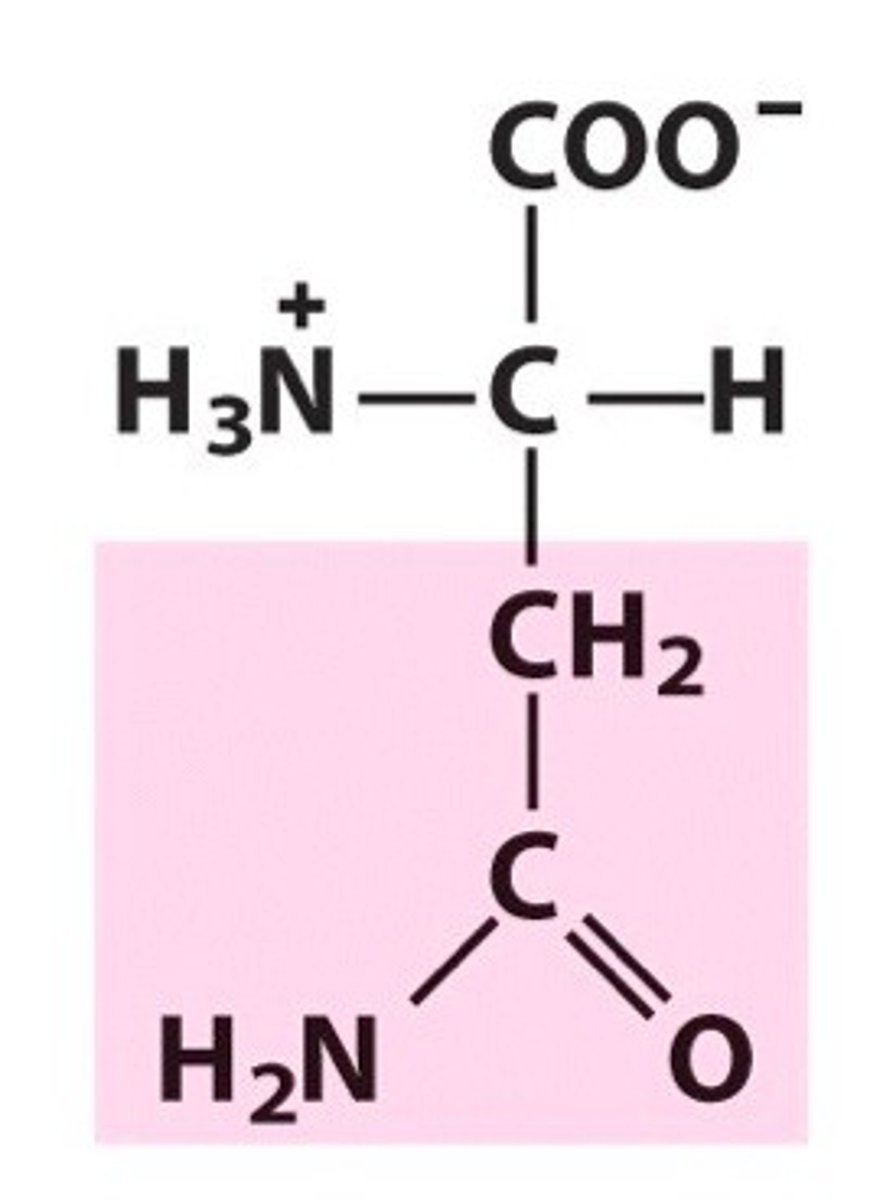
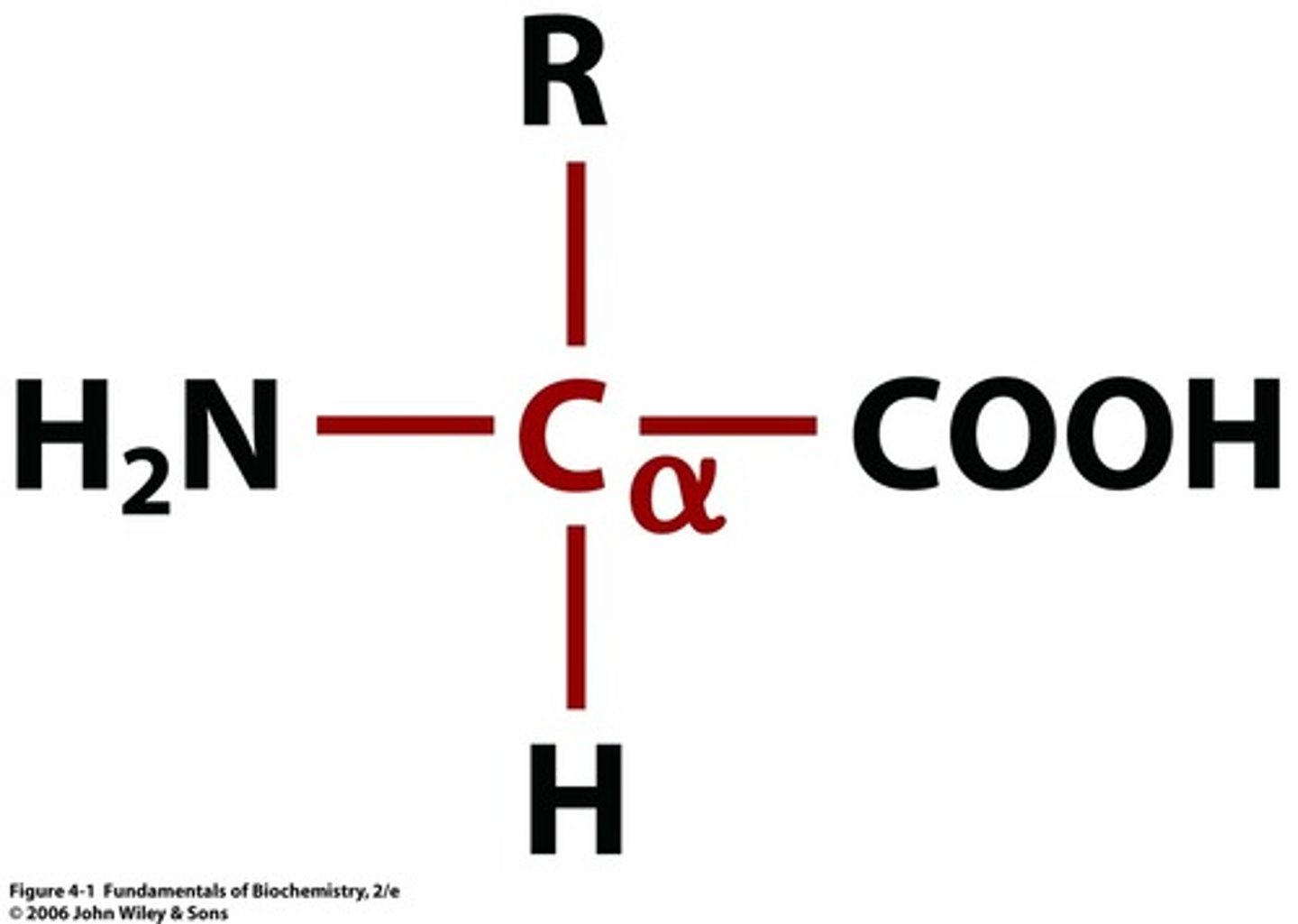
amino acid structure normal + charged in water
normal: central/alpha carbon connected to H, amine group (NH2), carboxylic acid (CO2H), and side chain
charged in water: central carbon connected to H, amine group (NH3, charged positive on nitrogen), carboxylic acid (CO2, charged negative on oxygen), and side chain
peptide bond
- very strong covalent bonds
- forms between carboxylic acid and amino group
- read amino acid left to right n-terminal (amino group) to c-terminal (carboxylic group)
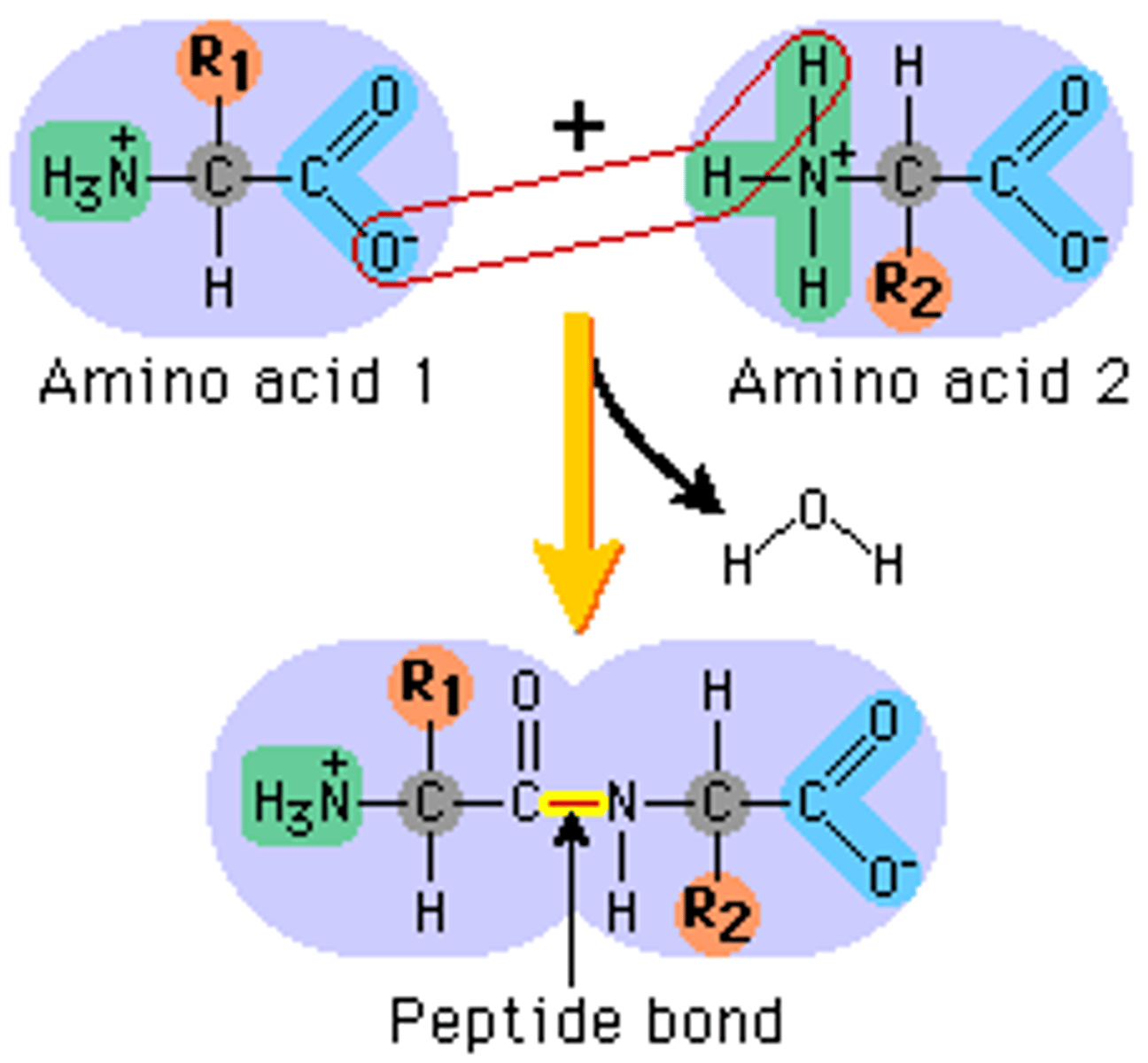
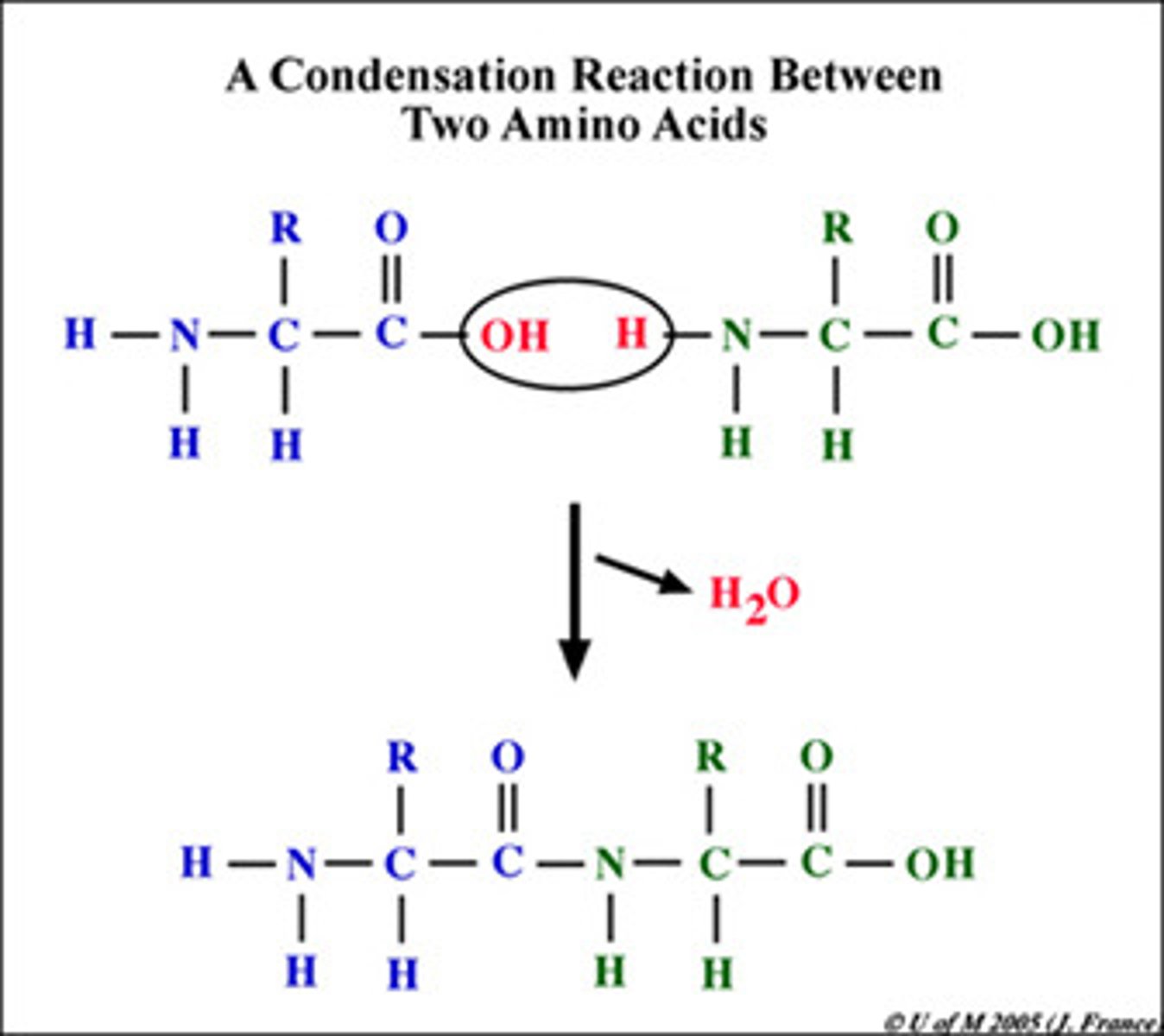
peptide bond formation
- condensation-dehydration reaction
- water released
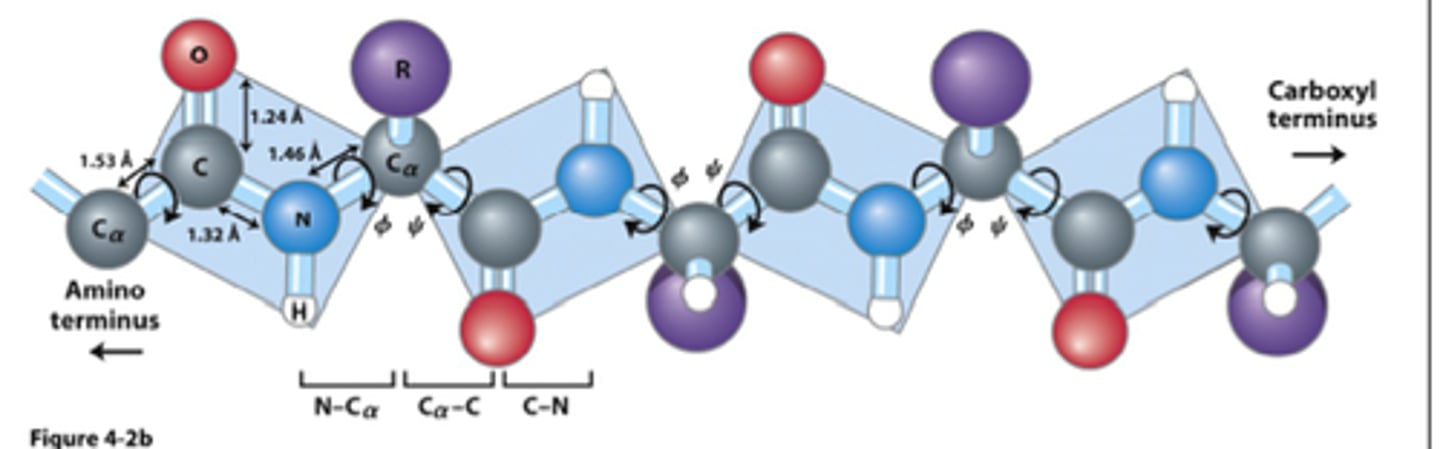
flexibility/rigidity of peptide bonds
- central carbon connected to amino/carboxylic group have very flexible bonds
- bonds between the 2 amino acids are very rigid
- covalent peptide bond is rigid = similar to double bond
- single bonds on either side of the alpha carbon are flexible + rotate to allow for folding of polypeptide into final protein conformation
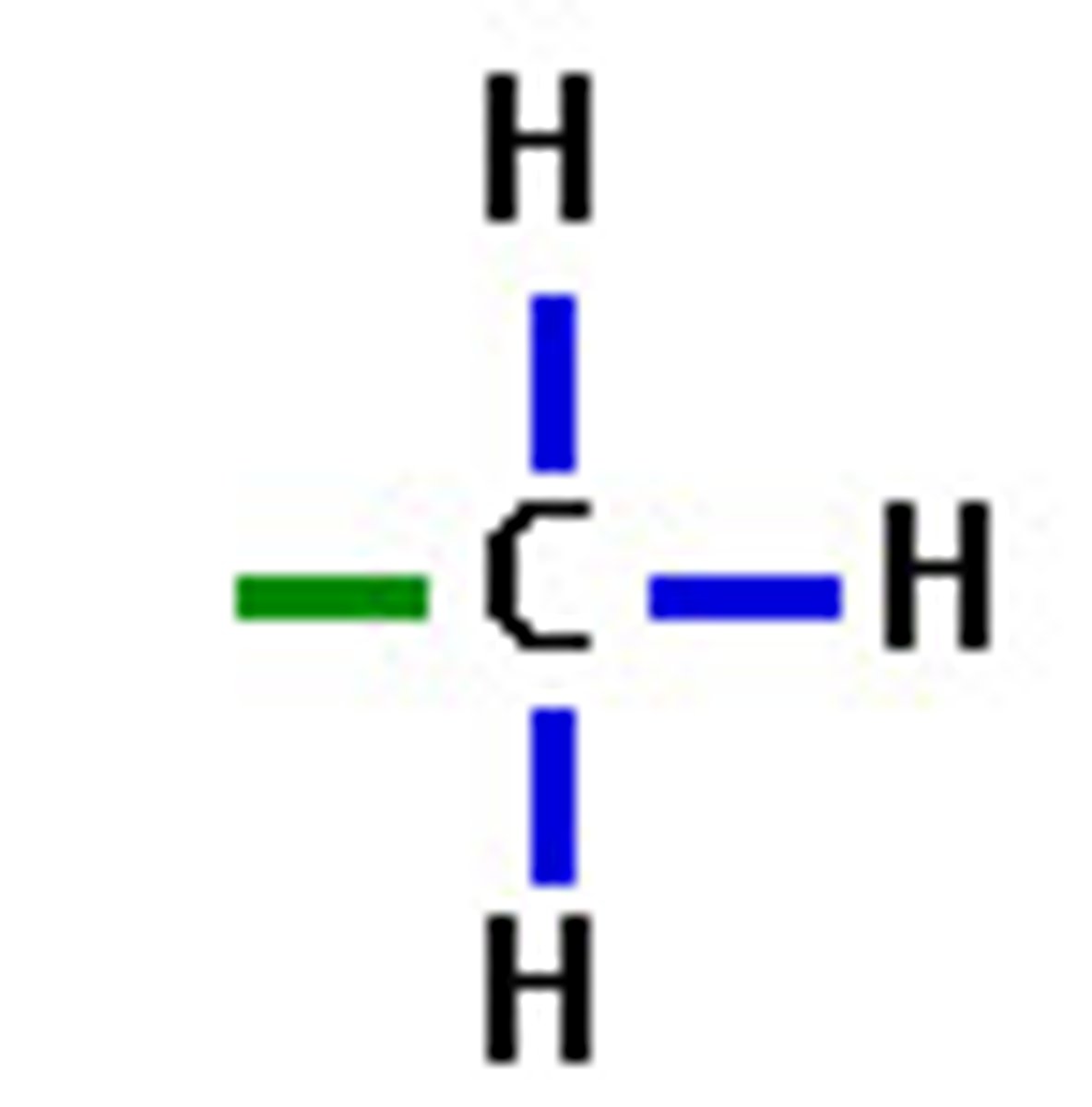
methyl functional group polarity
R-CH3
nonpolar
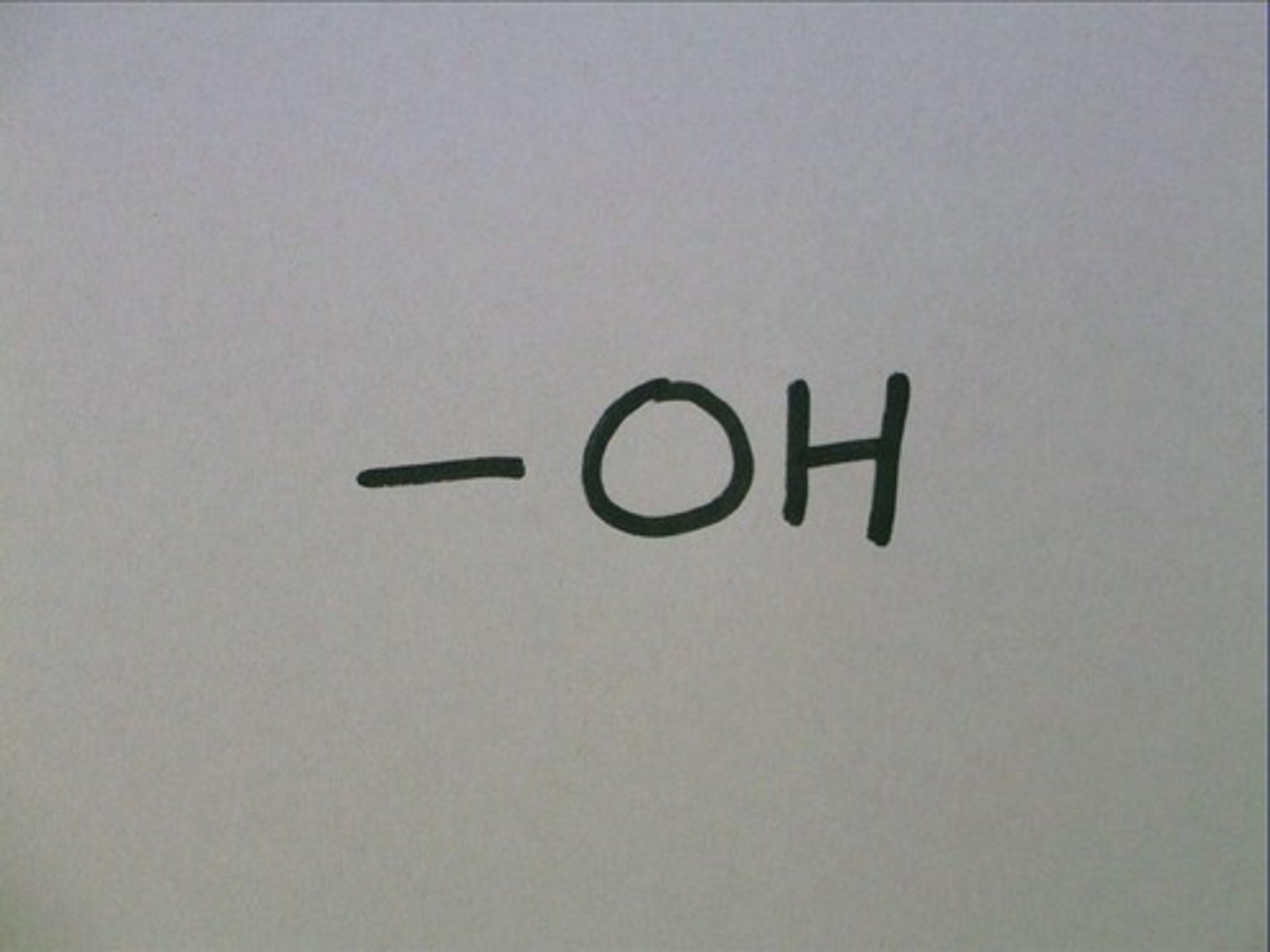
hydroxyl functional group polarity
R-OH
polar
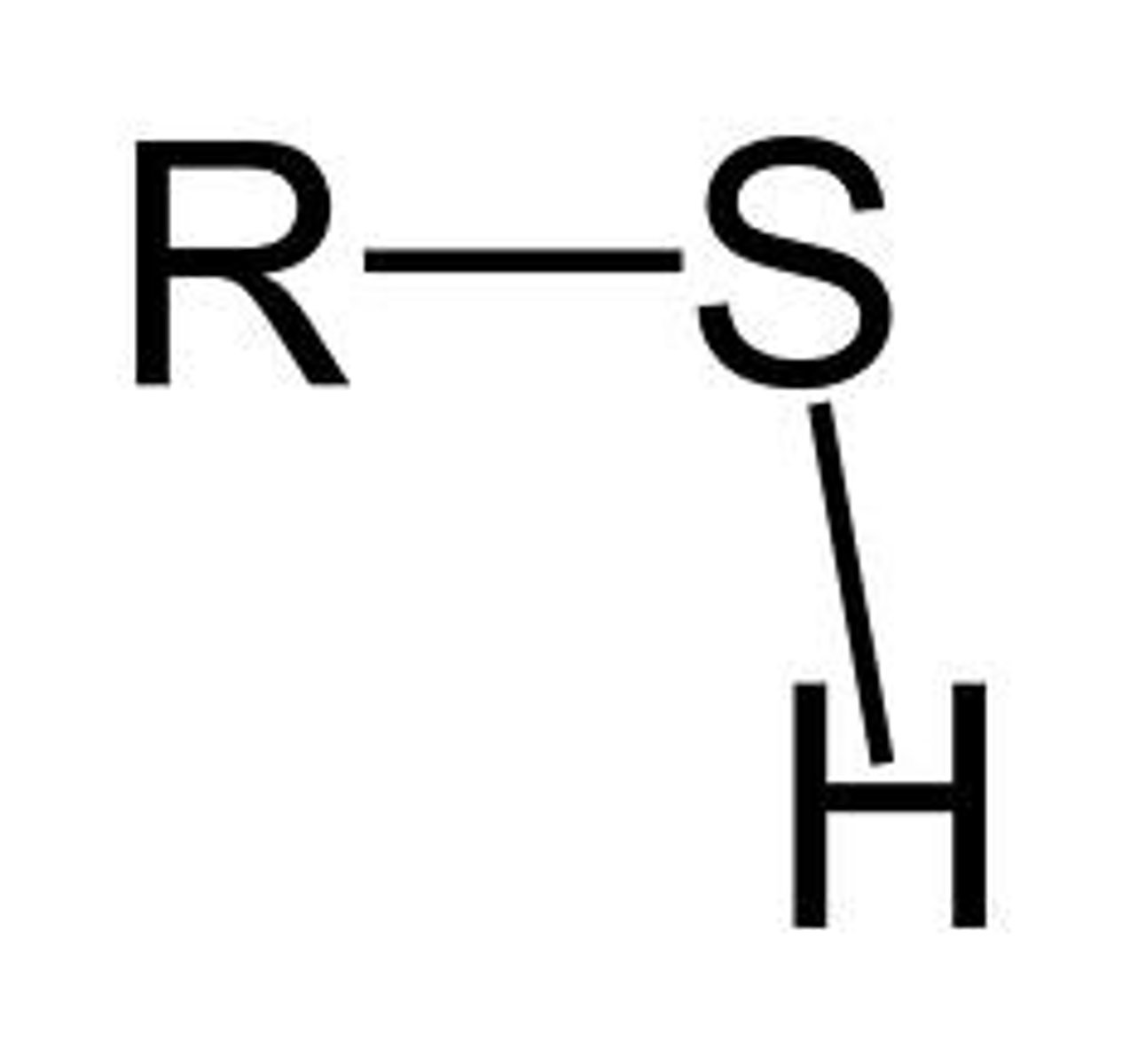
sulfhydryl functional group polarity
R-SH
polar
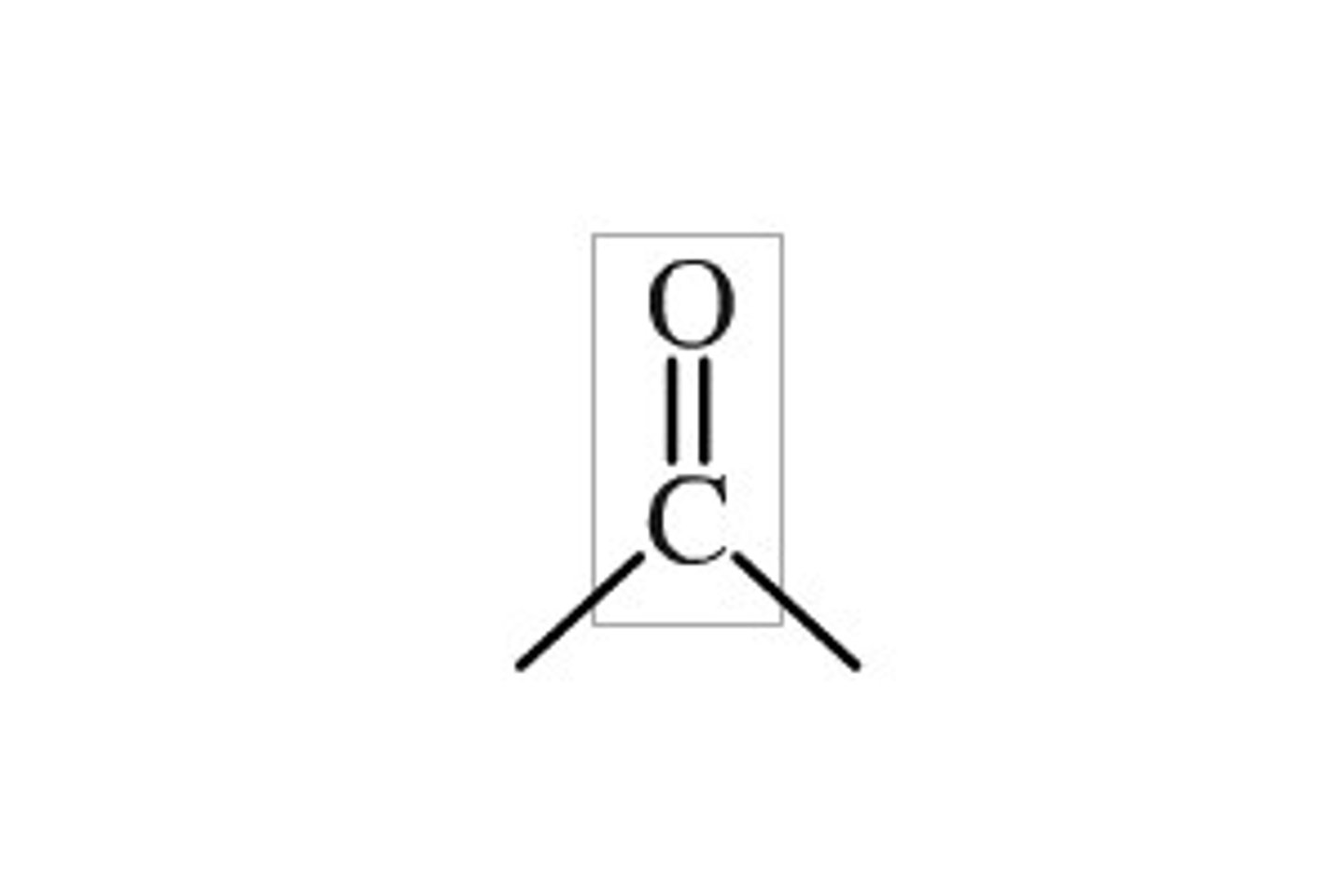
carbonyl functional group polarity
C=O
polar
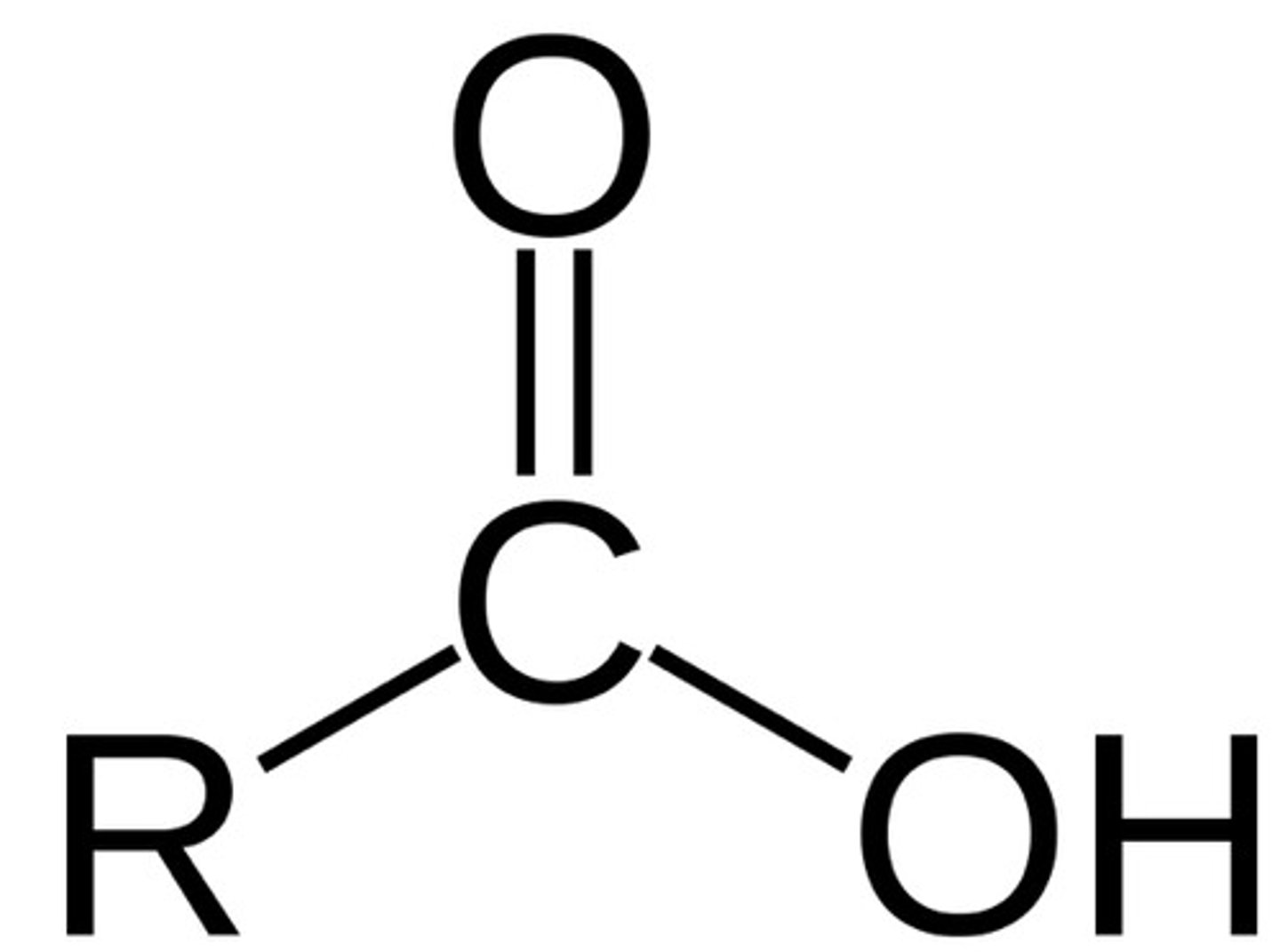
carboxyl functional group acid/base
acid
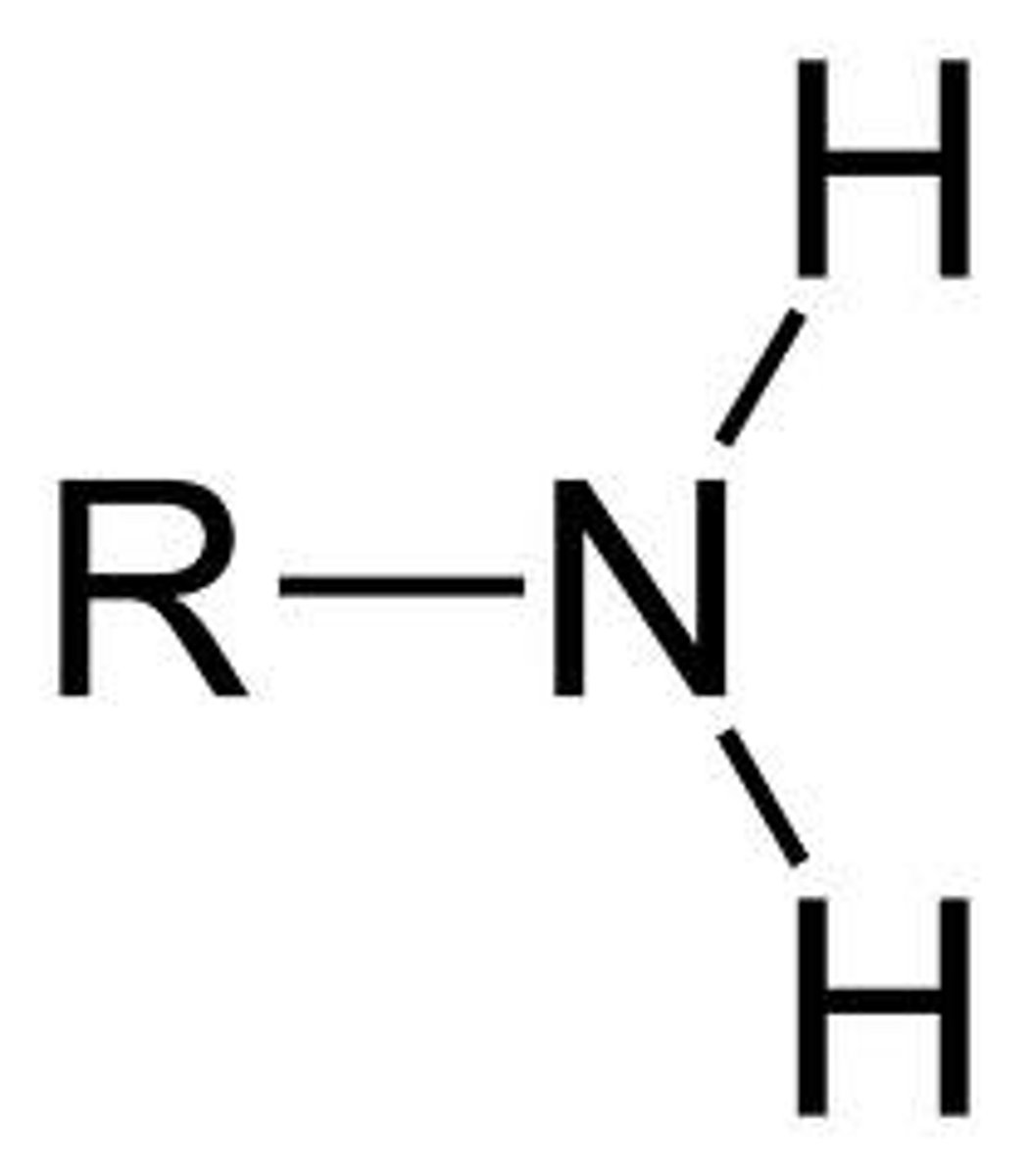
amine functional group acid/base
base
amide functional group polarity
polar
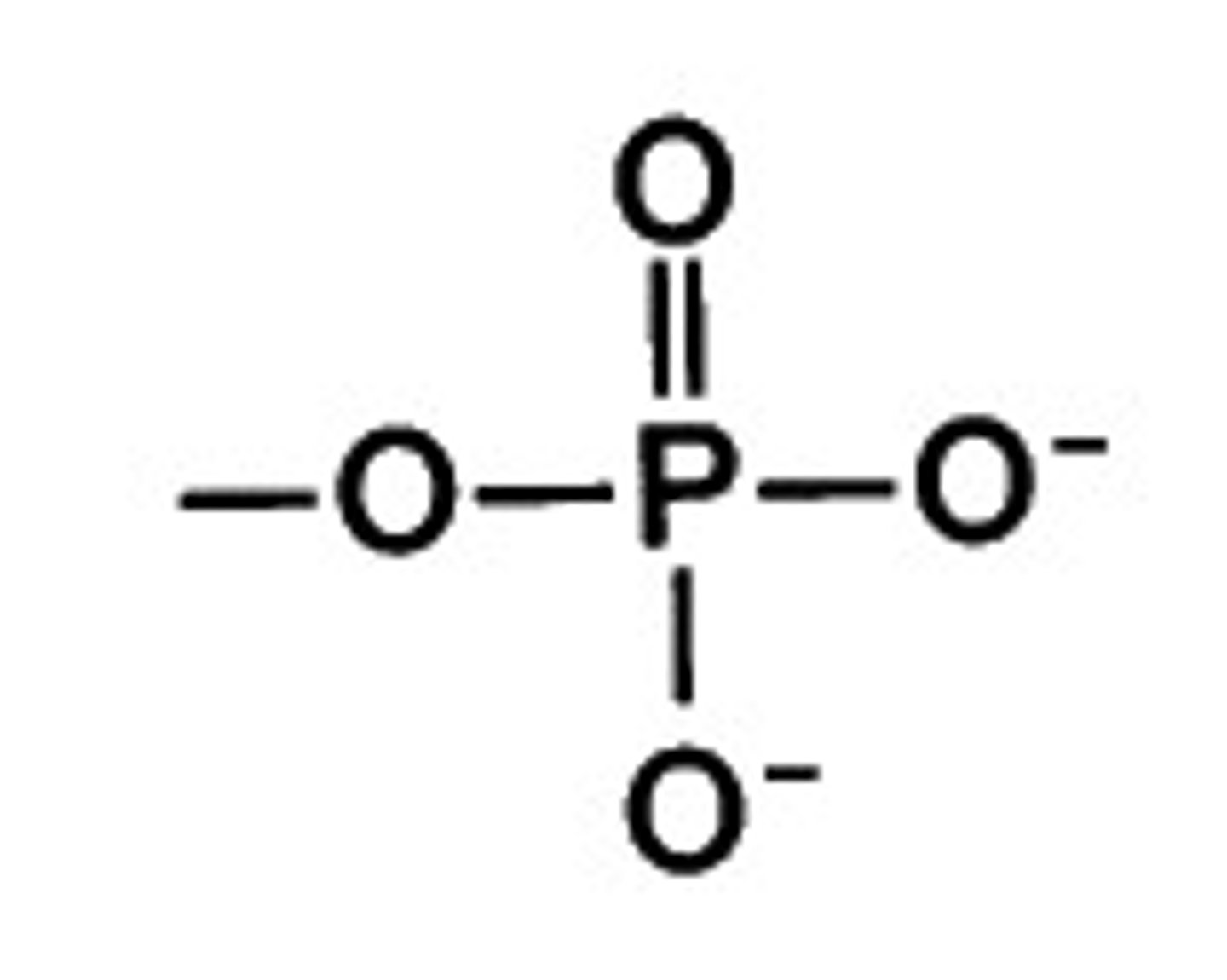
phosphate functional group acid/base
acid
translation
- converting the nucleotides in mRNA into amino acids to form proteins
- translating the language of 3-base codons into the language of amino acids
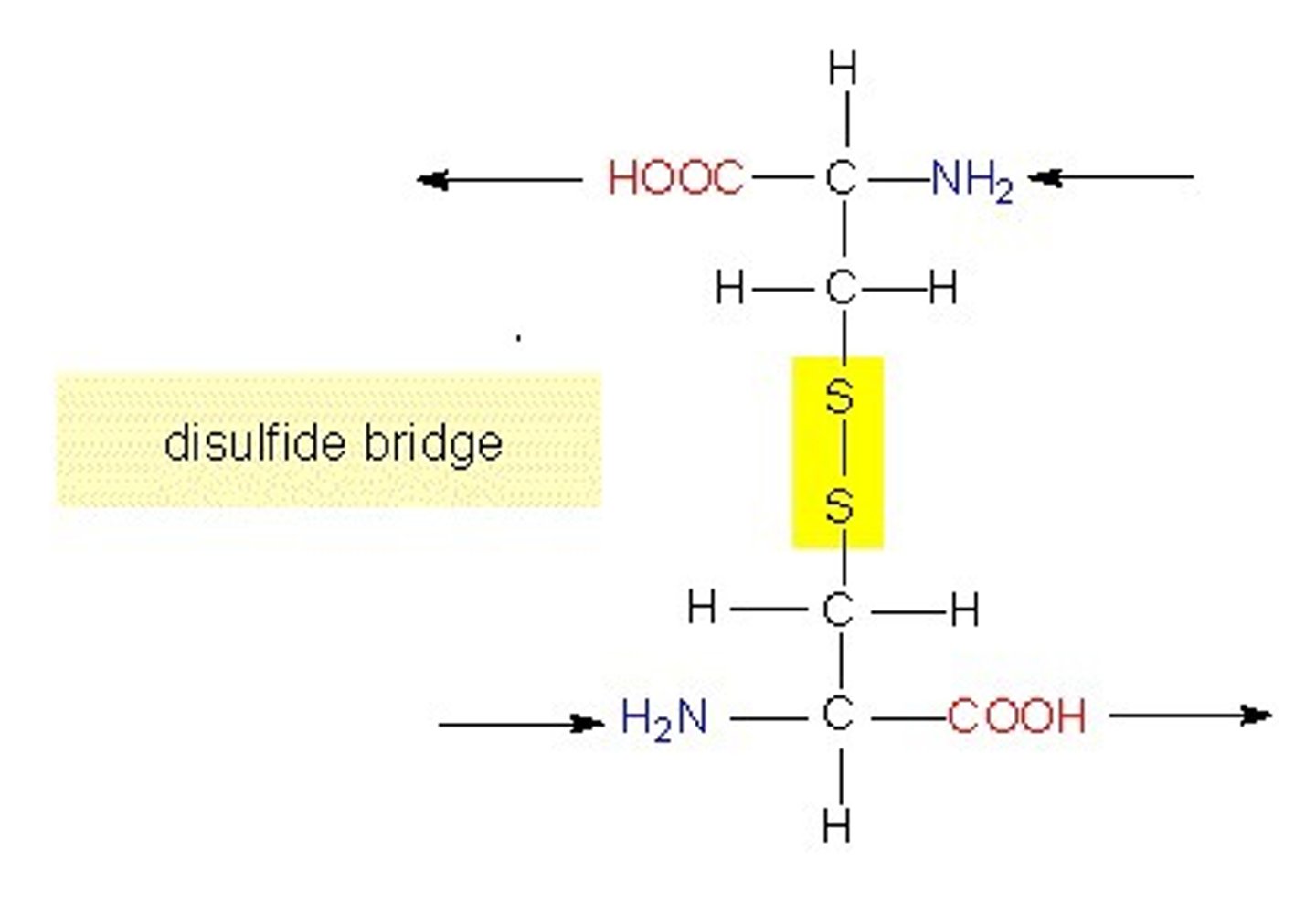
cysteine
- can form disulfide bridges --> important for tertiary structure
- oxidation of sulfhydryl groups --> electrons are lost
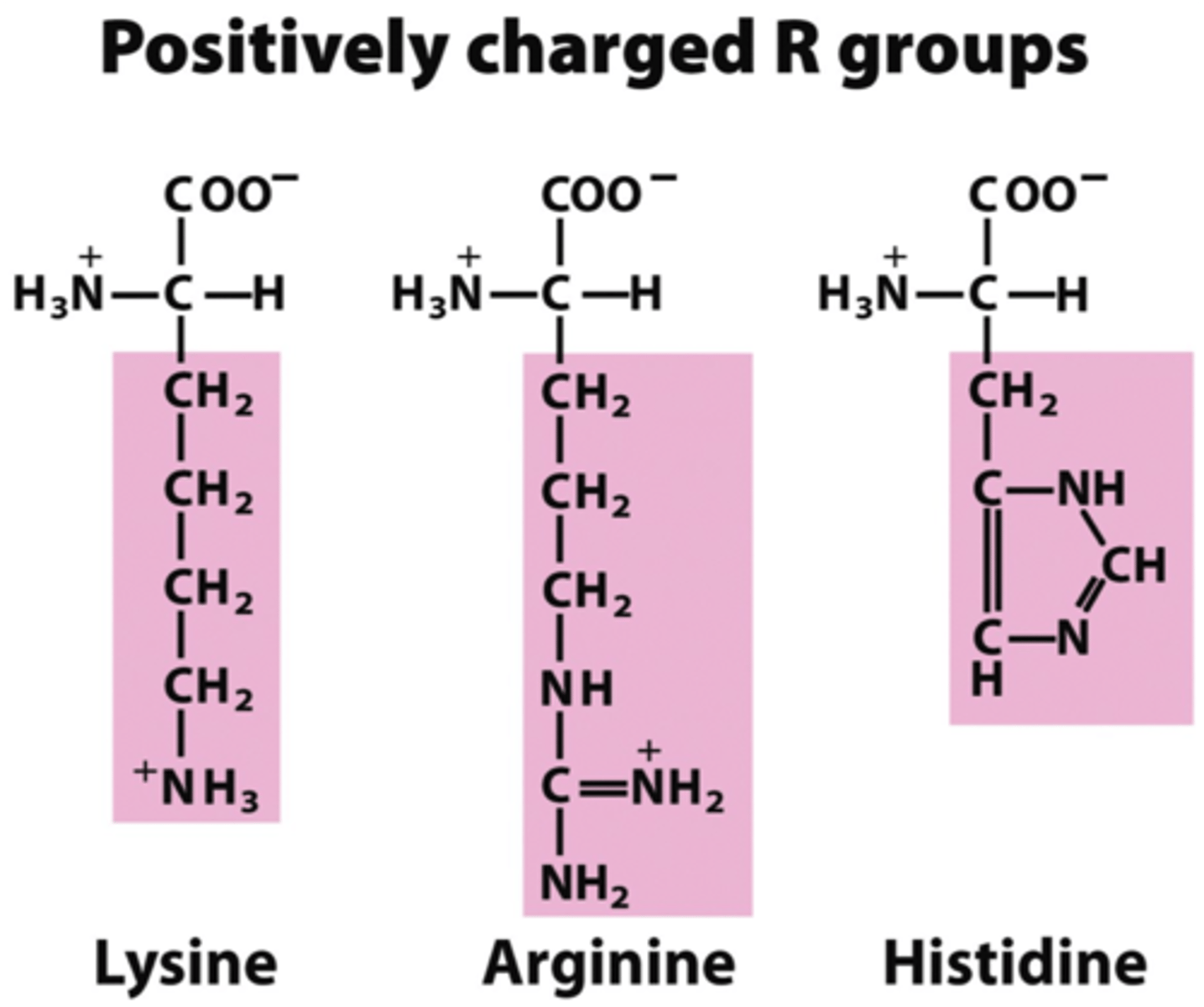
charged amino acids
Contain R groups with a negative or a positive charge
Form ionic bonds between negatively and positively charged amino acids
Hydrophilic
polar R groups
- atoms w/ high electronegativities (O,N,S) = R group is polar and hydrophilic
how to know if R group is an acid
carboxylic group
Carbon double bonded to oxygen and single bonded to OH --> will drop that H
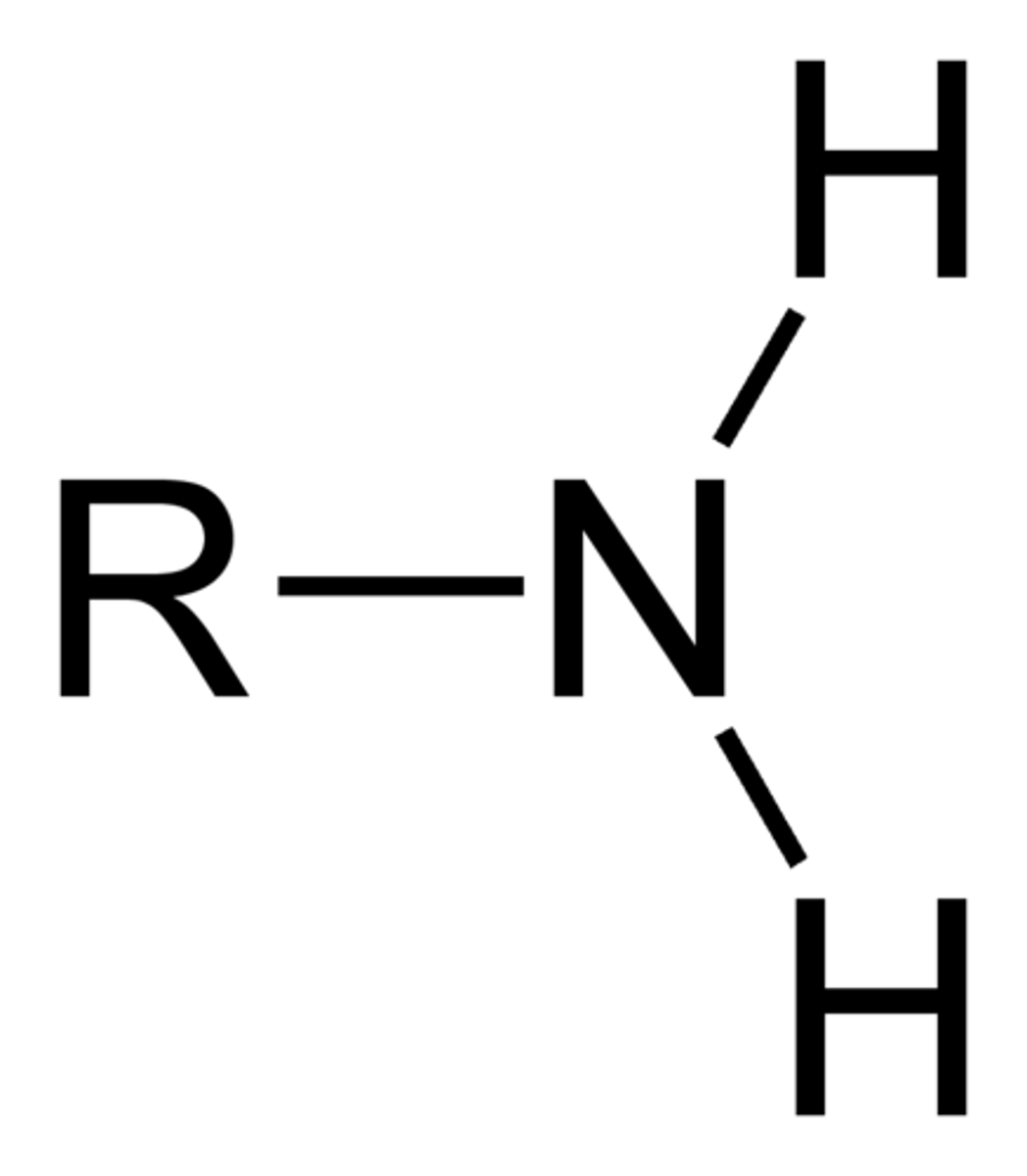
how to know if R group is a base
NH2 --> will want to gain an H
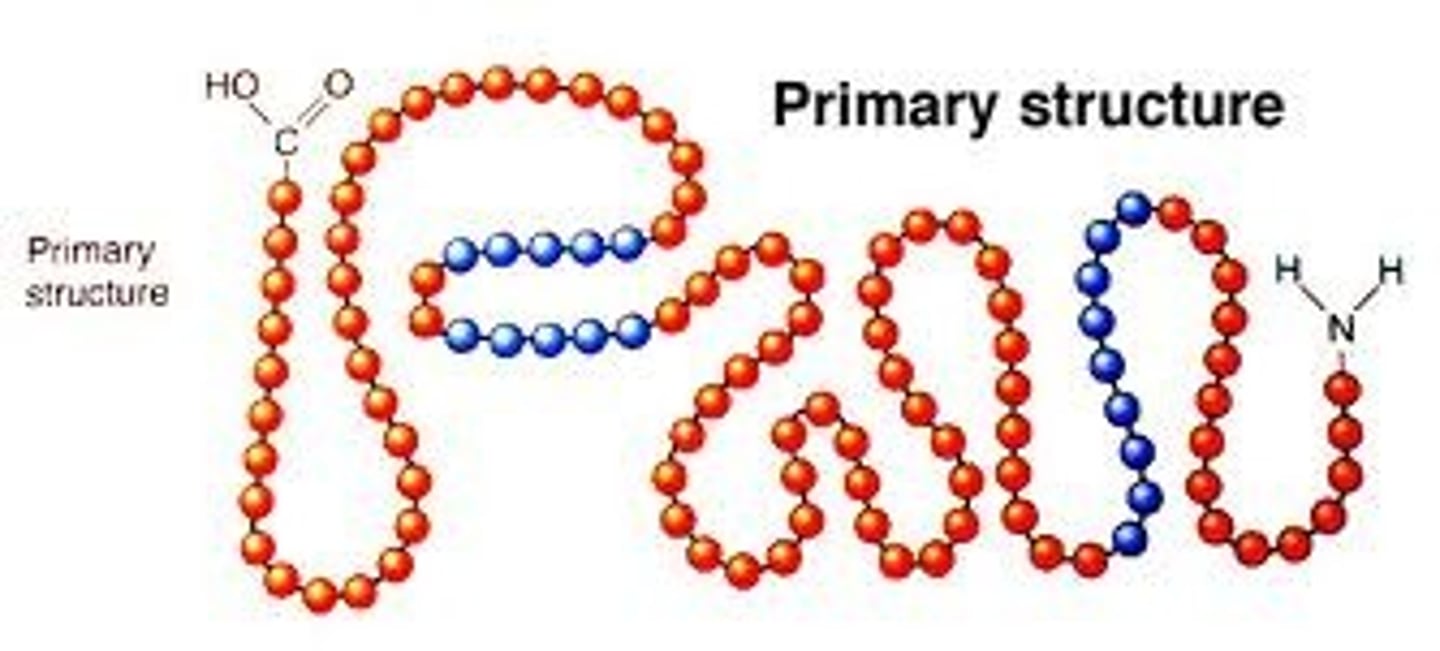
protein primary structure
- sequence of amino acids
- NH2 is the N terminus and the C=O - OH is the C-terminus
- always written N to C
- held together by covalent bonds (peptide bonds)
protein secondary structure
- alpha helix + beta pleated sheets
- held together by hydrogen bonding (no R groups involved)
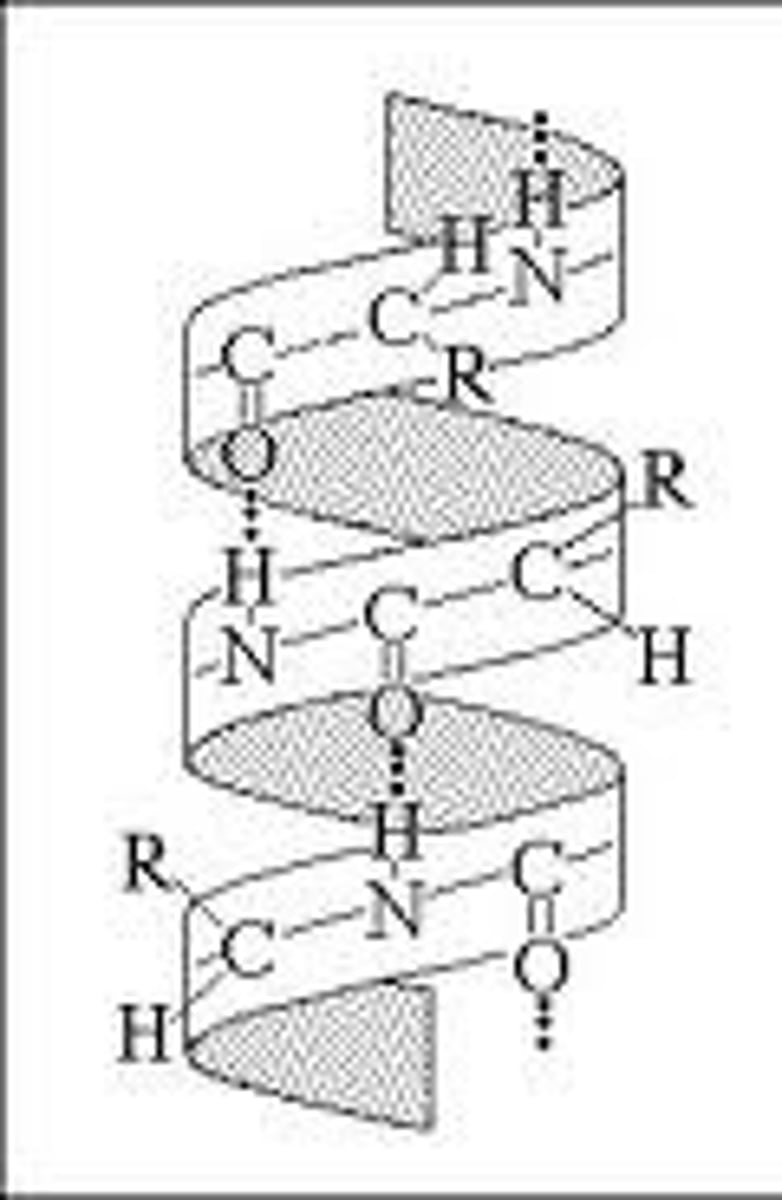
alpha helix
- hydrogen bonding between H and O = main stabilizing force
- hydrogen bonds every 4th peptide bond, linking C=O of one peptide bond to the N-H of another
- single polypeptide chain twists around on itself = rigid cylinder
- R groups stick out from the helix
- certain amino acids more likely to be found in alpha helix: Met, Ala, Leu, Glu, Lys (MALEK) = 3.6 AA per turn
beta pleated sheet
- hydrogen bonds main stabilizing force
- can have more than 2 strands, single beta sheet is not energetically favorable
- each side of the sheet may have distinct properties bc side chains alternately project above + below the plane of the sheet
- loops between each beta sheet provide flexible hinge regions
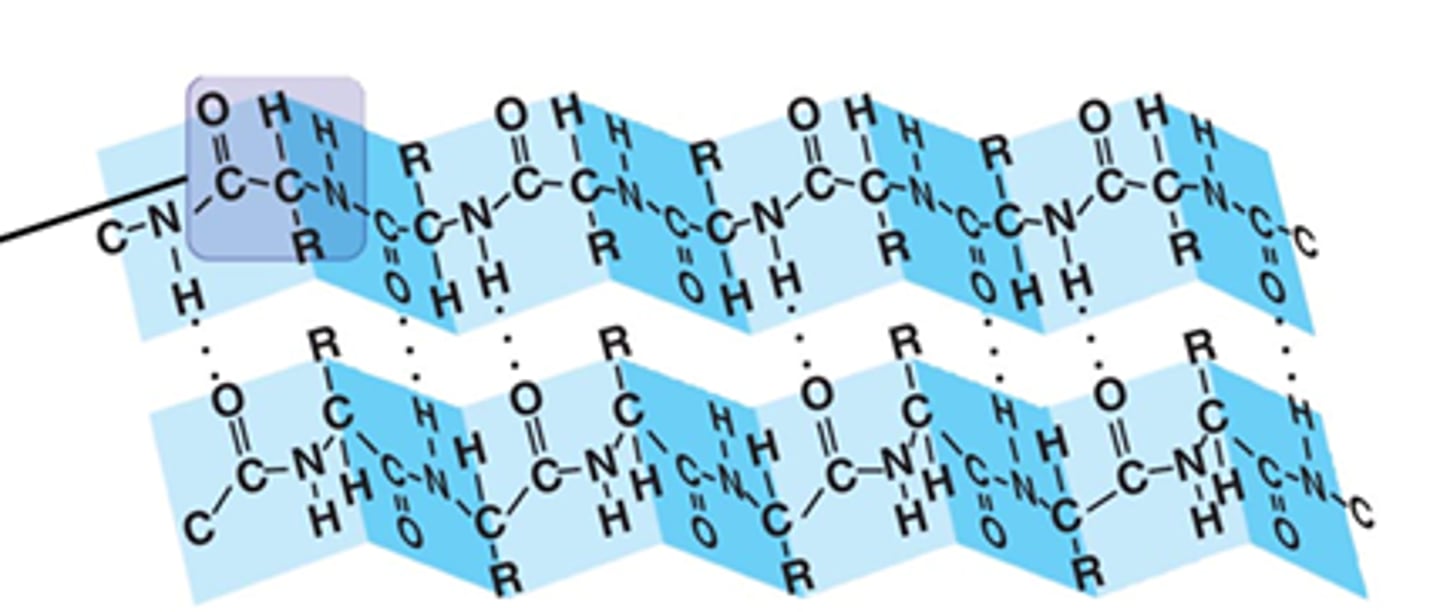
beta sheets running parallel or antiparallel
parallel: N-H and C=O groups can't line up as closely, hydrogen bonds are weaker + angled + farther apart
antiparallel: stronger, hydrogen bonds very close and straight
protein tertiary structure
- folded 3D structure
- hydrogen bonding involving R-groups, ionic bonds, hydrophobic interactions, disulfide bridges
protein quaternary structure
- multiple subunits to make a larger protein (is not for all proteins)
- subunits can be same (homo) or be diff (hetero)
- hydrogen bonding involving R-groups, ionic bonds, hydrophobic interactions, disulfide bridges = same types of non-covalent interactions that hold tertiary structure
which interactions that support tertiary/quaternary structures are covalent and non-covalent
non-covalent:
hydrogen bonds, ionic bonds, Van der Waals attractions, hydrophobic effect
covalent interactions:
disulfide bridges
protein tertiary - ionic bonds
- 2 FULLY charged ions attracted to each other
protein tertiary - hydrogen bonds
- form between partial charge in peptide backbone (N-H and C=O) or polar R groups
protein tertiary - van der waals attractions
- broader term for all types of weak intermolecular attractions between molecules, nonpolar side chains
- is the weakest
what happens to the diff types of bonds in protein tertiary structure when in water
- covalent: stay same
- ionic: decrease
- hydrogen: decrease
- van der waals: stay same
hydrophobic effect
- the tendency of nonpolar regions to aggregate in an aqueous solution + be shielded from water
polypeptide chains that span lipid bilayer
- usually alpha helix
- nonpolar amino acid side chains project outward to interact w/ hydrophobic membrane environment
disulfide bridges
- covalent bonds between cysteine residues on same or diff peptide chain
- form by the oxidation (lose electrons) of their -SH groups