Chapter 8 - Microbial Genetics
1/50
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
51 Terms
Gene expression
genetic information is used within a cell to produce the proteins needed for the cell to function
Gene Recombination
Genetic information can be transferred between cells of the same generation.
DNA replication
Genetic information can be transferred between generations of cells.
Genetics
The study of genes, how they carry information, how information is expressed and how genes are replicated
Chromosomes
structures containing DNA that physically carry hereditary information, the chromosomes contain genes
Genes
segments of DNA that encode functional products, usually proteins
Genome
all the genetic information in a cell
Genetic Code
A set of rules that determines how a nucleotide sequence is converted to an amino acid sequence of a protein
Central Dogma
DNA → RNA → protein
Transcription
DNA → mRNA
Gene expression
A gene is expressed when we have the protein product
In microbes, most proteins are either enzymatic or structural
Transcription
DNA → mRNA
scribe → copy
staying in the same language of nucleic acid
DNA
deoxyribonucleic acid (DNA)
Hydrogen bonds between the bases
strands are complementary
uniform width
antiparallel
order of the nitrogen-containing bases forms the genetic instructions of the organism
RNA
single stranded
many different types
5-carbon ribose sugar
extra OH group
contains uracil instead of thymine
Transcription - general
synthesis of a complementary mRNA strand from a DNA template
transcription begins when RNA polymerase binds to the promoter sequence on DNA
does not transcribed
proceeds in the 5’-3’ direction; only 1 of the two DNA strands is transcribed
transcription stops when it reaches the terminator sequence on DNA
3 stages of transcription
Initiation: RNA polymerase binds to a promoter
Elongation: Synthesis by adding complementary nucleotides
Termination: RNA polymerase reaches the terminator
Transcription
RNA polymerase bind to the promoter, and DNA unwinds at the beginning of a gene
RNA is synthesized by complementary base pairing of free nucleotides with the nucleotide bases on the template strand of DNA
the site of synthesis moves along DNA; DNA that has been transcribed rewinds
Transcription reaches the terminator
RNA and RNA polymerase are released and the DNA helix re-forms.
the strand that the mRNA uses → template strand as the mRNA uses that strand as a template and makes bases complimentary to it
the other strand is called the coding strand because it has the same nucleotides as the mRNA except thymine instead of uracil
Transcription - Eukaryotic Specific
transcription occurs in the nucleus whereas translation occurs in the cytoplasm
Exons are regions of DNA that code for proteins
Introns are regions of DNA that do not code for proteins
Spliceosome → a large RNA-protein complex that removes introns and splices together exons
composed of small nuclear ribonucleoproteins (snRNA) and RNA
RNA processing
In the nucleus, a gene composed of exons and introns is transcribed to RNA by RNA polymerase
Processing involves snRNPs in the nucleus to remove the intron-derived RNA and splice together the exon-derived RNA into mRNA
After further modification, the mature mRNA travels to the cytoplasm, where it directs protein synthesis.
Translation direction
mRNA → protein → translate between two languages (nucleic acid to amino acid)
The components required for protein synthesis
messenger RNA (mRNA): DNA → info → ribosomes
Ribosome
ribosomal RNA (rRNA): integral part of ribosomes
Ribosomal proteins
Amino acids
Transfer RNA (tRNA): transports amino acids during protein synthesis
Translation
mRNA is translated into the “language” of proteins
Codons are groups of 3 mRNA nucleotides that code for a particular amino acid
translation of mRNA begins at the start codon: AUG
Translation ends at nonsense codons: UAA, UAG, and UGA
Codons of mRNA are “read” sequentially
tRNA molecules transport the required amino acids to the ribosome
tRNA molecules also have an anticodon that base-pairs with the codon
amino acids are joined by peptide bonds
In bacteria, translation can begin before transcription is complete
The genetic code
61 sense codons encode the 20 amino acids
The genetic code involves degeneracy, meaning each amino acids is coded by several codons
The regulation of bacterial gene expression
constitutive genes are expressed at a fixed rate
other genes are expressed only as needed
inducible genes
repressible genes
Pre-transcriptional control → Induction
An inducible operon includes genes that are in the “off” mode with the repressor bound to the DNA, and is turned “on” by the environmental inducer.
When turned “on”, induction turns on gene expression
initiated by an inducer, which binds to the repressor, turning it inactive
The default position of an inducible gene is off
Pre-transcriptional control - repression
Repression inhibits gene expression and decreases enzyme synthesis
mediated by repressors, proteins that block transcription
Repressible operon default: “on” mode → meaning the DNA gene is being expressed because the repressor is inactive
turned “off” by the environmental corepressor and repressor.
Operon Model of Gene Expression
Promoter: segment of DNA where RNA polymerase initates (or promotes) transcription of structural genes
Operator: segment of DNA that controls transcription of structural genes
Operon: set of operator and promoter sites and the structural genes they control
unique to prokaryotes
In an inducible operon, structural genes are not transcribed unless an inducer is present
E.coli → enzymes of the lac operon are needed to metabolize lactose
In the absence of lactose, → repressor binds to the operator, preventing transcription
in the presence of lactose, the metabolite of lactose (allolactose → inducer) binds to the repressor
The repressor cannot bind to the operator, and transcription occurs
In repressible operons, structural genes are transcribed until they are turned off
Excess tryptophan is a corepressor that binds and activates the repressor to bind to the operator, stopping tryptophan synthesis
Lac operon - an inducible operon
Structure of the operon
promoter → operator → ZYA structural genes
operon is regulated by the product of the reg. gene (gene before promoter)
Lac operon → Repressor active
I gene is transcribed and translated to make a repressor protein
transcription → makes repressor mRNA
translation → makes active repressor protein
The active repressor protein binds to the operator region of the operon
When the repressor is bound to the operator → RNA polymerase can’t move forward to transcribe the structural genes (Y,Z, and A)
as a result → transcription is blocked and the genes that normally make the enzymes
Structure of the Lac Operon
The lac operon controls the breakdown of lactose in E. coli
includes 3 main region:
regulatory gene: makes the repressor protein, which blocks transcription
control region: contains promoter and operator
The promoter is where RNA polymerase binds to start transcription
The operator is the “switch” that the repressor binds to, turning the operon off
Structural Genes (ZYA): code for enzymes that break down lactose
lacZ: makes β-galactosidase (breaks lactose into glucose + galactose)
lacY: makes permease (helps lactose enter the cell)
lacA: makes transacetylase (detoxifies byproducts)
When the inducer (allolactose) binds to the repressor protein, the inactivated repressor can no longer block transcription. The structural genes are transcribed, ultimately leading to the production of the enzymes required for lactose catabolism.
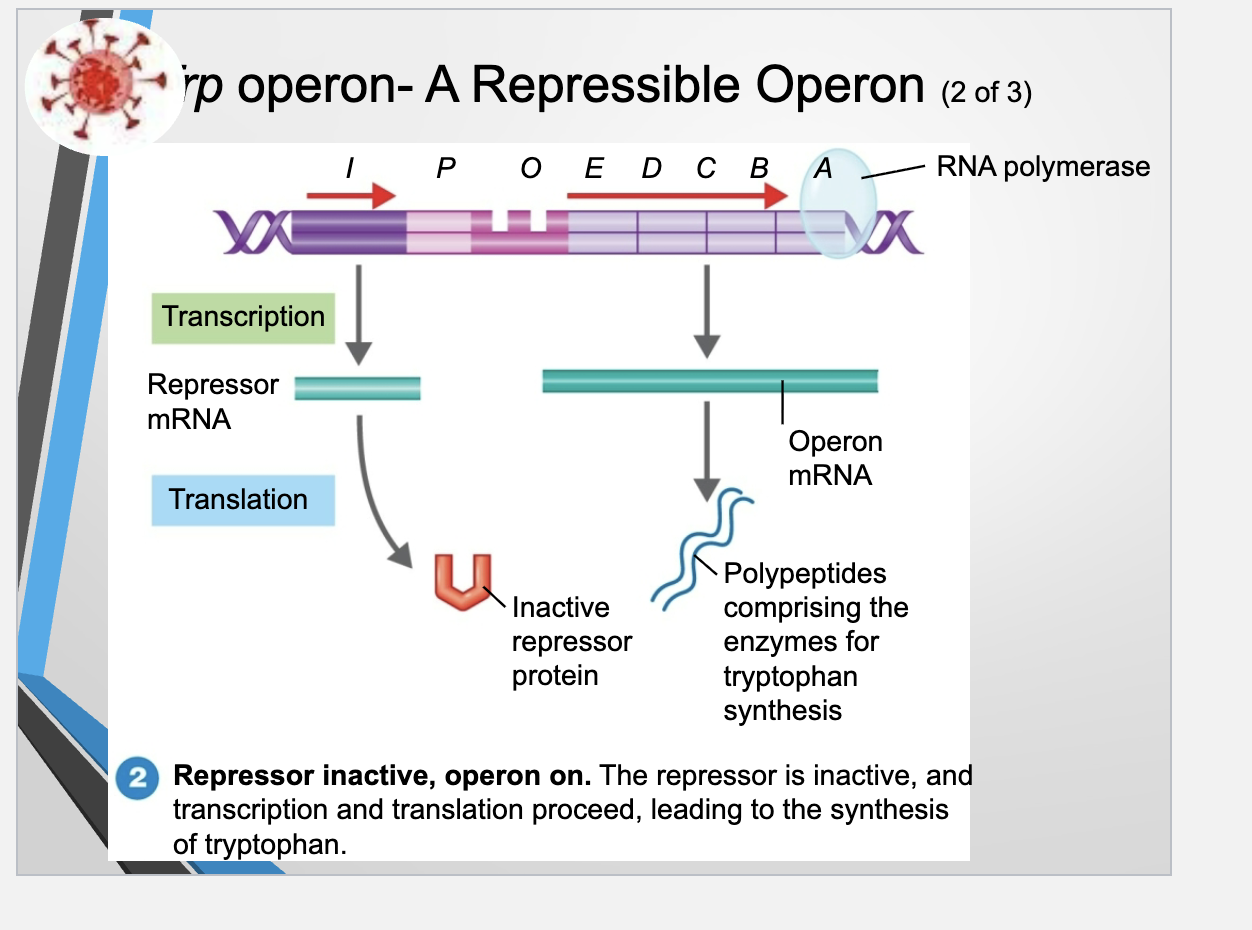
Trp operon - A Repressible Operon
The operon consists of the promoter and operator and structural genes that code for the protein
the operon is regulated by the product of the regulatory gene
The Trp operon controls the production of enzymes needed to make the amino acid tryptophan
The regulatory gene → repressor mRNA → inactive repressor protein (cannot bind to the operator region) → allows RNA polymerase to attach to the promoter and trancribe the structural genes → resulting mRNA is then translated into enzymes that synthesize tryptophan
the cell produces tryptophan when its levels are low
When tryptophan levels are high:
the amino acids acts as a corepressor
tryptophan binds to the inactive repressor protein → changing its shape and activating it
the active repressor then binds to the operator region of the DNA
blocks RNA polymerase from binding to the promoter or moving forward → transcription stops
no mRNA or enzymes are made
Summary: Repressor active → operon off.
The operon shuts down when enough tryptophan is present — a negative feedback loop that prevents waste of energy and resources.
Changes in genetic material
Mutation: a permanent change in the base sequence of DNA
mutations may be neutral, beneficial or harmful
Mutagens are agents that cause mutations
Spontaneous Mutations occur in the absence of a mutagen
Types of mutations
Base Substitution
point mutation
change in 1 base of DNA
can be deleterious or result in no change
CG → AT
Missense Mutation
Base substitution results in a change in an amino acid
Nonsense Mutation
base substitution results in a nonsense (stop) codon
The stop codon is premature, resulting in a truncated protein product '
Frameshift mutation
insertion or deletion of one or more nucleotide pairs
shifts the translational reading frame
Chemical Mutagens
increase the mutation rate
Nitrous Acid: causes A to bind with C instead of T
Nucleoside analog: incorporates into DNA in place of a normal base, causes mistakes in base pairing
antiviral medications
Frameshift mutagens
Radiation
ionizing radiation (x-rays and gamma rays) causes the formation of ions that can oxidize nucleotides and break the sugar-phosphate backbone of DNA
non-ionizing radiation (UV light) produces thymine dimers
Photolyases
enzymes that are found in some microorganisms that use light energy to break thymine dimers
Say if A pairs with G instead of T, doing so → the DNA strand would be cut along with the neighbors surrounding the mispair → the missing patch would be replaced with correct nucleotides → DNA ligase seals the gap
Frequency of mutations
spontaneous mutation rate = 1 in 109 replicated base pairs or 1 in 106 replicated genes
The mutation rate is dependent on the organism in question
Mutations occur randomly along the genome
Mutagens increase the mutation rate by 10-5 or 10-3 replicated gene
Identifying Mutants
Positive (direct) selection detects mutant cells because they grow or appear different than unmutated cells
Negative (indirect) selection detects mutant cells that cannot grow or perform a certain function
use of replica plating
Auxotroph: mutation that has a nutritional requirement that was absent in the parent
Replica Plating process
A sterile velvet surface is pressed on the grown colonies in a master plate
Cells from each colony are transferred from the velvet to NEW plates (one with histidine and the other one without)
plates are incubated
Growth on plates is compared. A colony that grows on the medium with histidine but cannot grow on the medium without histidine is auxotrophic for histidine
Ames test
exposes mutant bacteria to mutagenic substances to measure the rate of reversal of the mutation
indicates the degree to which a substance is mutagenic
2 cultures of Salmonella that lost the ability to synthesize histidine are prepared
The suspected chemical mutagen is added to the experimental sample only, and the control stays without it, but rat liver is added to both
Each sample is poured into a medium that does not contain histidine → Incubation → Only bacteria that were histidine dependent reverted into having histidine colonies. → shown to be more on the plate that had the suspect mutagen than the one that did not
The higher the concentration of mutagen used, the more revertant colonies are obtained
DNA and chromosomes
eukaryotic DNA is linear and segmented
Bacteria usually have a single circular chromosome made of DNA and associated proteins
DNA replication - ORI
The origin of replication is where replication begins
One strand serves as a template for the production of the second strand
semiconservative replication
DNA replication of a plasmid
Most bacterial DNA replication is bidirectional
Each offspring cell receives one complete copy of the DNA molecule
Replication is highly accurate due to the proofreading capability of DNA polymerase
Mutation: 1 in every 10 billion base incorporate
Enzymes in DNA replication
DNA gyrase: relaxes supercoiling ahead of the replication fork
DNA ligase: makes covalent bonds to join DNA strands, Okazaki fragments and new segments in excision repair
DNA polymerases: synthesize DNA, proofread and facilitate repair of DNA
Helicase: unwinds double stranded DNA
Methylase: adds methyl group to selected bases in newly made DNA
Primase: an RNA polymerase that makes RNA primers from a DNA template
Topoisomerase or gyrase: relaxes supercoilng ahead of the replication fork; separates DNA circles at the end of the DNA replication
DNA replication process
Topoisomerase and gyrase relax the strands
targeted by quinolone antibiotics
helicase separates the strands
A replication fork is created
Primase lays down the RNA primer of which the new strand is synthesized
DNA polymerase adds nucleotides to the DNA strand in the 5 ‘ to 3 ‘ direction
The leading strand is synthesized continuously (5 prime to 3 prime), and the lagging strand is synthesized discontinuously w Okazaki fragments
DNA polymerase removes RNA primers, Okazaki fragments are joined with the help of the DNA polymerase and ligase
Genetic Transfer
Horizontal gene transfer: transfer of genes between cells of the same generation
only for prokaryotes
Vertical gene transfer: transfer of genes from an organism to its offspring
Mobile genetic elements
move from 1 chromosome to another
transposons (both prokaryotes and eukaryotes)
Or move from 1 cell to another
plasmids (prokaryotes only)
Plamids
self-replicating circular pieces of DNA
1 to 5% of the size of a bacterial chromosome
Often code of proteins that enhance the pathogenicity of a bacterium
Conjugative plasmid:
Type of plasmid that can move from one cell to another through carrying transfer or tra genes that code for proteins intended to carry a sex pilus → bridge like structure between cells
Dissimilation plasmids
encode enzymes/genes for the breakdown or to metabolize unusual organic compounds such as pesticides, hydrocarbons or aromatic compounds
Resistance factors
encode antibotic resistance
Transposons
segments of DNA that can move from 1 region of DNA to another
Enzyme recognizes and binds to the inverted repeat sequences at both ends of the transposon
Enzymes and transposons form a looped structure called the transposition complex
The enzyme cut the transposon out of the donor DNA molecule
It finds a new target site somewhere else in the DNA
It is inserted into a new location → joining the DNA sequence with the IR at both ends
Horizontal gene transfer in bacteria
conjugation
Bacterial plasmids can be transferred from 1 cell to another via a sex pilus or a bridge
if F+ goes to F-, F- cell becomes F+
Transformation
genes are transferred from 1 bacterium to another as “naked” DNA
the “naked” DNA can be incorporated into the bacterial genome
for stable integration into the bacterial genome, crossing over must occur
review Griffith’s experiment
Transduction
DNA is transferred from 1 donor cell to a recipient via a bacteriophage
Generalized transduction: random bacterial DNA is packaged inside a phage and transferred to a recipient cell
A phage infects the donor bacterial cell
phage DNA and proteins are made, and the bacterial chromosome is broken into pieces
pieces of bacterial DNA are packaged into a phage capsid
donor cell lyses and releases phage particles containing bacterial DNA
phage infects the recipient cell
Inside the recipient cell, the donor bacterial DNA can combine (recombine) with the recipient’s own DNA.
This creates a recombinant cell, meaning its genome is now a mix of both the donor and recipient bacteria.