TRANSCRIPTION
1/14
Earn XP
Description and Tags
Exam 1
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
15 Terms
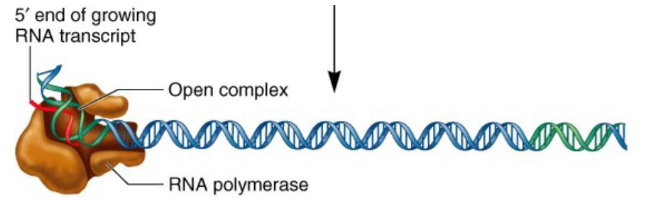
RNA polymerase
An enzyme which reads and transcribes DNA and synthesizes a single strand of RNA using the DNA template strand. It knows where to start transcribing the RNA using promoter regions within the DNA sequence.
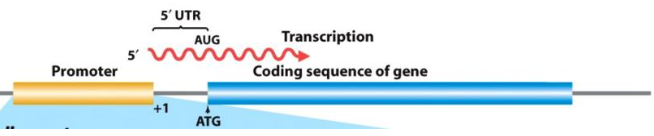
Promoter
A region encoded within DNA indicating where transcription should start. Always in front of the gene that needs to be transcribed.
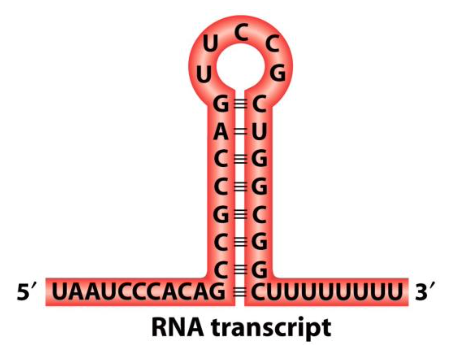
Terminator
A region (hairpin) encoded within DNA indicating transcription to stop. A signal which causes RNA polymerase to fall off from the DNA, and a completed strand or RNA.
In some bacteria species there are two methods of termination; rho dependent which uses a protein rho which slides along the RNA transcript, and rho independent which does not use rho and relies on the hairpin and weak A-U bonds to break apart.
Sigma factor
Associated with RNA polymerase, scans for the promoter region.
Abortive initiation
Because RNA polymerase and the sigma factor are so tightly bound to the promoter, for RNA polymerase to separate for transcription small RNA fragments are made until polymerase breaks free from the promoter.
TFIID
Recognizes the TATA box and other DNA sequences near the transcript start point.
TFIIH
Unwinds DNA at the transcript start point, and releases RNA polymerase from the promoter (acts like helicase).
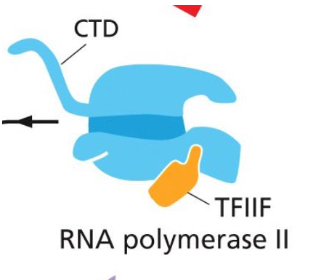
C-terminal domain (CTD)
The tail of RNA polymerase which goes through physical changes that let RNA polymerase move away from the promoter.
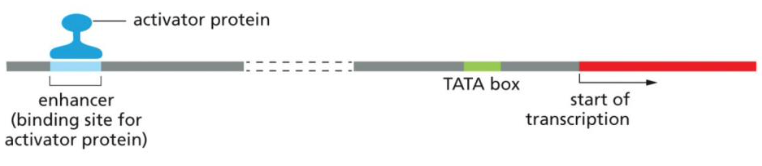
Enhancers
A binding site along the DNA strand for activator proteins.
Activator proteins
Regulatory proteins which increase the rate of transcription of a given gene.
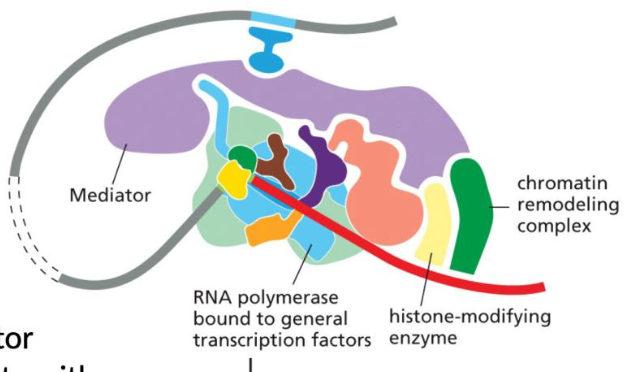
Mediator
A complex which allows activator proteins to communicate with RNA polymerase 2 and the general transcription factors.
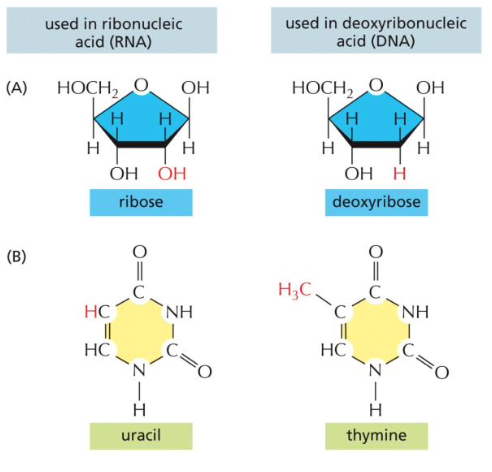
How does RNA differ from DNA?
RNA has an OH group, ribose where DNA has an H group, deoxyribose. As opposed to thymine (T) in DNA, RNA uses uracil (U). RNA is single stranded.
How are transcription and replication similar/different?
RNA does not need a primer and is less accurate. It is okay for RNA to make mistakes because we are mainly focused on DNA and the genes it will encode. RNA also reads from 5’ to 3’.
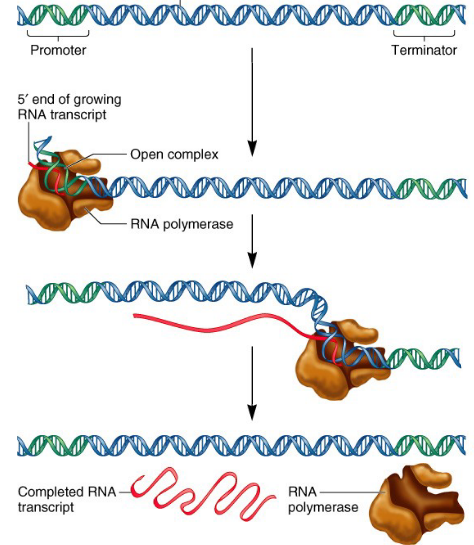
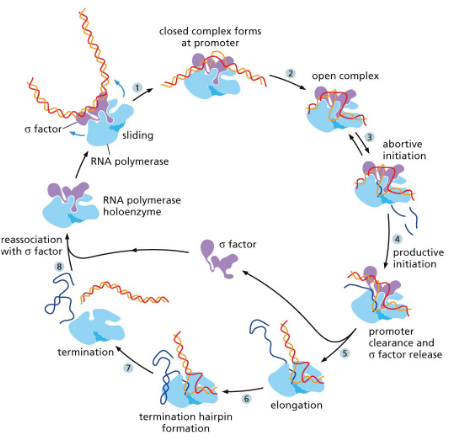
Steps of transcription in bacteria:
INITIATION:
Begins with RNA polymerase holoenzyme which consists of the RNA polymerase core enzyme and a sigma factor which scans for a promoter region that it can recognize and bind to.
When the promoter is found the holoenzyme binds tightly to the DNA (sigma factor recognizes bases) and opens up the DNA helix (forms 10 bp long region of unpaired DNA). Abortive initiation (removing RNA polymerase holoenzyme from promoter) occurs and makes little RNA fragments until the complex falls off from the promoter.
ELONGATION:
Elongation proceeds as normal until the terminator sequence in DNA is reached.
TERMINATION:
Contain multiple U’s and A’s (weakest bonds) in the form of a hairpin, which leads to full dissociation of complex and RNA transcript.
Steps of transcription in eukaryotes:
INITIATION:
Has 3 types of RNA polymerase, but most important is RNA polymerase II which transcribes most genes including all that encode proteins (RNA polymerase I and III transcribe genes encoding tRNA, rRNA and various small RNAs),
Requires many additional proteins called general transcription factors that help position RNA polymerase at the promoter, aid in pulling apart the DNA strands, and release RNA polymerase from the promoter to start elongation. Most important are TFIID and TFIIH.
Has a CTD (C-terminal domain) which is the “tail” of RNA polymerase that undergoes changes that allow RNA pol to move away from promoter.
Requires the mediator protein complex, which allows activator proteins to communicate to RNA pol II and general transcription factors.
ELONGATION:
Tightly coupled with RNA processing. There are 3 steps, addition of the 5’ cap, RNA splicing and the addition of the 3’ poly-A tail.
The 5’ cap is made when RNA processing proteins are carried on the tail (CTD) of RNA pol. As RNA is synthesized, the capping proteins from the tail begin to process the 5’ end.
RNA splicing is the removal of intron sequences from newly synthesized RNA, but there is also alternative splicing where one gene can be spliced many different ways which produces different, related proteins.
For the addition of the 3’ poly-A tail, the end of the mRNA molecule is encoded within the DNA sequence, so when RNA pol transcribes these sequences they are recognized by two proteins; the cleavage stimulation factor and the cleavage and polyadenylation specificity factor. When these factors bind additional proteins come in to cut the RNA, and poly-A polymerase comes in to synthesize the poly-A tail which does not need a template.