GENETICS 3
1/174
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
175 Terms
What is the difference betwen constitutive and regulated transcription?
constituative transcription is always on and always transcribing genes. Regulated transcription are only expressed under certain environmental conditions
What is one advantage and one disadvantage of regulating gene expression?
pros
not metabollically costly → genes only expressed when needed
responds to environmental conditions
cons
needs to accurately respond to enviornment
not instant response
What is a repressor protein? What is its allosteric site?
A repressor protein binds to specific sites on DNA to inhibit transcription of nearby genes. Its allosteric site allows it to change shape in response to ligand binding, affecting its binding to DNA.
What is an inducer? What is a corepressor?
a molecule that initiates gene expression by binding to a repressor protein, causing it to detach from the DNA
A corepressor is a molecule that binds to a repressor to enhance its ability to repress gene expression.
What is an activator protein?
proteins that have a DNA binding proteins. They block transcription initiation and occupy the same space where RNA pol. would normally bind or prevent formation of the open promoter complex.
What is an allosteric effector? What is an allosteric inhibitor?
allosteric effectors allow binding and allosteric inhibitors prevent binding
What are operons? What are two advantages of operons?
Operons are groups of genes and their shared regulatory region. They allow efficient transcriptional responses to the environment and enable coordinated regulation of functionally related genes.
What conditions trigger expression of the lac operon? What conditions prevent expression in the lac operon?
if both glucose is present and lactose is present, the Lac operon is on. However, if glucose is present, lactose is absent, or cAMP levels are low, the lac operon is turned off.
What is a polycistronic mRNA?
three genes transcribed together: Permease (lacY), Beta-galactosidase (b-gal) and transacetylase (lacA)
In the lac operon: What is the operator? What binds it? What is the consequence of this? When is it bound?
the lac operon contains an operator region where the repressor protein binds. This binding prevents RNA polymerase from transcribing the downstream genes. It is bound when lactose is absent, inhibiting gene expression.
In the lac operon, what binds the repressor protein and what is the consequence of this?
The repressor protein binds to the operator, preventing RNA polymerase from transcribing the downstream genes. This occurs when lactose is absent, keeping the operon off.
How is allolactose formed? How does lactose enter the cell if the operon is off?
It can be produced from B-gal acting on lactose and can also be broken down into glucose and galactose by B-gal. If alloctose is not present, the repressor will bind the operator and block transcription initittion.
If the operon is off, shouldn’t there be no β-gal in the cell, leading to no allolactose being produced? Gene expression is never completely zero. there is always some low level of permease and B-gal present.
When does cAMP bind CAP? What is the consequence of this?
in the absence of glucose, the concentration in the cell of cyclic AMP (cAMP) increases. This is caused by activaton of adenylate cyclase, which converts ATP to cAMP. When glucose is present, this enzyme does not catalyze the reaction.
cAMP binds to CAP causing it to bind to DNA at the CAP binding site. this facilitates RNA polymerase binding and activates transcription.
What is the difference between basal and leaky expression?
leaxy expression arises as a consequence of repressor binding being reversible and not 100% efficient → low amount of transcription
Basal expression arises as a consequence of a lack of expression and activation.
What types of mutations lead to constitutive mutants? Which act in cis? Which act in trans?
Mutations in operators and I- mutations that alter DNA binding domains are constitutive mutants.
Operator mutations are cis-acting, only influencing transcription of genes on the same chromosome, and I-mutations are trans-acting.
What types of mutations lead to non-inducible mutants? Do they act in cis or in trans?
super repressor mutations make the operon unresponsive to induction by allolactose. Mutations in I that alter its allosteric domain without altering its DNA binding properties. Even when allolactse is present, the protein cannot release the DNA. Is mustations act in trans
Define negative feedback
a change in a certain direction triggers responses that counteract that change, helping to maintain homeostasis.
Contrast typical characteristics of catabolic and anabolic operons.
Catabolic operons like the lac operon are often
inducible by the presence of a particular nutrient.
Anabolic operons are typically repressible by the
end product (negative feedback)
What is attenuation? What is an advantage of attenuation compared to a simple on/off mechanism?
attenuation allows the magnitude of expression to be tightly controlled in response to various concentrations of the endproduct. An advantage is, attenuation acts as a dimmer switch rather than just an on/off switch, providing finer regulation of gene expression.
What is the function of the 1-2 stem loop?
1,2 stem loop forms when a ribosome does not quickly associate with the nascent trpL mRNA. The 1,2 stem loop slows transcription to allow for this binding
What determines whether a 2-3 or a 3-4 stem loop will occur?
The formation of the 2-3 or 3-4 stem loop is determined by the concentrations of the amino acids, influencing whether the ribosome pauses at the attenuator region during transcription. When the ribosome stalls, a 2-3 stem loop is formed. When the ribosome progresses quickly, the 3-4 stem loops is formed
Suppose the trp operon had its attenuator codons mutated to stop codons, what would be the expected result?
the ribosome would stop early instead of stalling. This allows the terminator hairpin to always form, so the operon is always attenuated → very low trp gene expression regardless of tryptophan levels.
Explain how alternative sigma factors can give rise to genome wide alterations in gene expression.
Each sigma factor recognizes its own promoter consensus sequence. When a cell switches which sigma factor is active, RNA polymerase binds that sigma factor and begins transcribing all genes that have its matching promoters. Because many genes share these promoter motifs, changing sigma factors instantly shifts transcription toward an entire regulon, producing genome-wide changes in gene expression.
Explain how translational regulation for ribosomal proteins functions. Is this negative feedback? Why or why not?
one of the products of each operon can bind their own mRNA at the shine-salgarno seq. preventing their translation. This provides negative feedback and is the main way production of these proteins is regulated. If we have a lot of proteins, we stop making more of them.
Explain how antisense RNAs can block translation.
if antisense RNA binds to an mRNA it can prevent its translation. Bacteria can tolerate low expression, but high expression can cause problems if genes are getting disrupted by the transposon. The gene has two promoters oriented in opposite directions. P-in is a weak promoter, this drives expression of the transposase. P-out promter is a strong promter and drives expression of an overlapping antisense RNA. When these RNAs base pair the shine-salgarno seq. and start codon are sequestered and cannot bind the ribosome
Compare and contrast tryptophan’s role as a co-repressor in the trp operon and GlcN6P’s role in regulating glmS. How are these mechanisms similar? How are they different?
Tryptophan acts as a co-repressor by binding to the trp repressor, enabling it to attach to the operator and inhibit transcription of the trp operon. In contrast, GlcN6P binds to the glmS mRNA and promotes its self-cleavage, thereby regulating its own synthesis. Both mechanisms provide feedback to regulate gene expression in response to metabolite levels, but they do so through different molecular interactions.
Give two reasons why gene regulation is more complicated in eukaryotes as compared to bacteria.
chromatin states can alter gene expression
regulatory sequences can be “long range” rather than just in the promoter
What two types of regulation are present within multicellular eukaryotes that are not present in bacteria? Explain what is meant by each
temporal (during development)
spatial (tissue specific)
Compare and contrast the characteristics of the core promoter with that of an enhancer sequence.
How are they similar and how are they different?
core promoter
region that is upstream of the transcription start site
binds general transcription factors and RNA pol II
often contains a TATA box
enhancers and silencers
enhancers increase the expression of target genes
Silencers down-regulate target genes
not constrained to be upstream of genes
can be intronic or exonic, close or far
enhancer/silencer mosules have many binding sites for different TFs
this allows them to integrate the activities of different transcription factors to produce different outputs
can turn genes on or off and can turn gene expression up or down
cis acting sequences bind trans-acting proteins
Explain how different patterns of SHH expression are accomplished in the brain and in the limbs
Different expression patterns are needed because they have different functions. There is a long-range enhancer that is limb-specific and there is a short range enhancer for the brain
different transcription factors present = different patterns of expression in different tissues
Propose a plausible outcome for using CRISPR to replace the cobra SHH enhancer with the mouse SHH enhancer within a cobra
took normal mouse limb specific enhancer and used crispr to replace it with the cobra genome. This caused a mouse to develop without limbs. This suggests that morphological evolution is largely driven by differential regulation of genes
Summarize the general argument for why morphological evolution is presumed to more often be driven by regulatory rather than coding changes. Illustrate this argument using the example of humans and chimps
regulatory mutations that do not alter coding sequences can cause large morphological changes .oolkit genes direct development by changing the timing and amount of these genes and drastic differences can arise. humans and chimps share ~99% of amino acid identitiy in protein sequence. genes are regulated in different amounts at different times during devleopment.
A mutation in the beta-globin LCR causes Thalassemia. Propose a plausible mechanism of action of this mutation.
The mutation in the beta-globin Locus Control Region (LCR) leads to decreased transcriptional activation of the beta-globin gene, resulting in reduced production of hemoglobin and the development of Thalassemia. This occurs because the LCR is crucial for enhancing the expression of the beta-globin gene.
Define modularity in the context of enhancers and silencers. Suppose that enhancers were not modular, how would this change the consequences of mutations in these regions?
modularity means that components of an enhancer module can be altered without altering the effects of other componenets. If enhancers were not modular, mutations in one part could dramatically affect the entire function of the enhancer, leading to more widespread and unpredictable effects on gene regulation.
Why are some non-coding enhancer elements so highly conserved?
Some non-coding enhancer elements are highly conserved because they play critical roles in regulating gene expression across different species, ensuring that essential developmental and physiological processes are maintained.
How can the property of modularity give rise to different species using different binding sites and different trans acting proteins to regulate a gene, but the pattern of gene expression remains similar between these species?
Modularity allows different enhancers to interact with various transcription factors, enabling diverse regulatory combinations while maintaining conserved gene expression patterns across species.
Explain the mechanism of activation of the Gal genes in yeast. Your answer should explain the roles of UASG Gal80, Gal4, and Gal3 both in the presence and in the absence of galactose
Gal1, Gal2, Gal7, and Gal10 genes are needed for galactose import and utilization as a carbon source. Each of these genes has its own promoter, but is regulated by the transcription factor, Gal4. Gal4 binds an enhancer element called an upstream activator sequence (UAS). Gal4 is constitutively expressed, but is bound to Gal80 (also constitutive) and cannot activate transcription in the absence of galactose.
in the absence of galactose, gal 3 is present in the cytoplasm. in the presence of galactose, gal3 moves to the nucleus, where it binds gal8- and causes the release of gal80 from gal4. This allows for activation of gal genes because the activation domain is no longer obscured
Explain the mechanism of repression of the Gal pathway in yeast. Your answer should explain the roles/location of Mig1 both in the presence and absence of glucose
Mig1 is made in the presence of glucose. It binds a silencer between UAS and the gal genes. This recruits another protein called Tup1, and this complex interferes with the Gal4 enhancer element.
Upon glucose depletion, Mig1 is phosphorylated and exported from the nucleus.
Compare and contrast the gal pathway with the lac operon. How are they similar and how are they different?
Both the gal pathway and the lac operon are regulatory systems controlling gene expression in response to sugar availability. They share mechanisms like the use of specific transcription factors (Gal4 for gal genes and LacI for lac operon) and the presence of enhancer or silencer elements. However, they differ in their sugars (galactose vs lactose), regulatory proteins (Gal80/Gal3 in the gal pathway vs LacI in the lac operon), and the differing responses to glucose availability.
How are enhancers directed to only act to increase the transcription of specific target genes?
Insulator sequences bind proteins and direct enhancers to interact with the intended promoter. This blocks communication between enhancers and other promoters. There can be a preference for one enhancer over another. Mutations in insulator sequences can yield inappropriate enhancer activation.
Identify each of the following histone marks as either generally heterochromatic or euchromatic: H3K27me3, H3K9ac, H3K9me3, H3K4me
H3K27me is a repressive mark in facultative heterochromatin.
H3K9me3 is a repressive mark in constitutive heterochromatin
K3K9ac and H3K4me are euchromatic marks
they alter chromatin structure
they are transmissible during cell division (mitotically stable)
they are reversible
they do not alter DNA sequence
Histone deacetylases remove acetyl groups from histones. Would we expect greater activity of these enzymes in a particular genomic region to increase or decrease transcriptional activity of that region and why?
Greater activity would decrease transcriptional activity because histone deacetylases promote tighter chromatin structure, leading to reduced gene expression.
Contrast nucleosome sliding with nucleosome ejection
nucleosome sliding: nucleosome remains bound but is displaced such that the regulatory sequences are exposed
nucleosome rejction: removal of the nucleosome and moving it to another sequence, exposing the regulatory sequence
Explain the regulation of PHO5 in yeast in as much detail as possible
PHO5 encodes a phosphatase that is repressed in the presence of excess phosphate. It is activated upon phosphate starvation.
PHO4 encodes a protein that facilitates regulation. Under high phosphate conditions, this protein is localized to the cytoplasm. under low phosphate conditions, this protein is localized to the nucleus.
In high phosphate conditions, the TATA box is not exposed and is blocked by the -1 nucleosome. UASp2 is not exposed and is blocked by the -2 nucleosome. UASP1 is bound by Pho2 (transcription activator). Pho2 is bound by NUA4, an acetylase. Promoter histones are not acetylated
In low phosphate conditions, Pho4 is present and binds Pho2. NUA4 acetylates nearby histones, opening the chromatin. Pho2/Pho4 complex displaces the -2 nucleosome, another molecule of Pho4 binds, and the SWI/SNF complex is recruited.
What are lncRNAs and what is their role in X inactivation?
IncRNAs are long noncoding RNAs. They play a key role in gene regulation, X inactivation. One gene is Xist, which is active on the inactivated chromosome and inactive on the activated chromosome. This gene encodes an lncRNA that coats only the X chromosome from which it was derived. This is its only function. This binding facilitates HDACS and methylases to the chromosome, condensing it to a Barr body and rendering it inactive
Define allele specific expression and genomic imprinting.
Allele-specific expression is the phenomenon where one of two parental alleles for a gene is expressed more or less than the other
Genome imprinting is when only the allele inherited from one parent is active while the other is silenced
Explain how IGF2 expression from exclusively the paternal allele is accomplished
On the paternal chromosome, ICR and H19 are methylated, and no insulator binds. Long-range enhancer acts on the IGF2 promoter; paternal IGF2 is expressed, but H19 is not.
If imprinting is disrupted, improper development occurs! altering appropriate gene dosage!
Explain the argument for sexual conflict in the evolution of maternal IGF2 imprinting in species with litter sizes greater than one and common multiple paternity
The system is likely a remnant of sexual conflict that arose due to male and female reproductive strategies being different.
IGF2 promotes the growth of the fetus - it promotes higher provisioning of maternal resources.
Males benefit most from having each of their offspring use as many maternal resources as possible.
Females benefit most from equal partitioning of resources to all of her offspring (and not provisioning offspring with too many resources.
Contrast the production of siRNAs and miRNAs
siRNAs are typically produced from long double-stranded RNA precursors and target specific mRNAs for degradation, while miRNAs are derived from single-stranded RNA transcripts that form hairpin structures, regulating gene expression by binding to complementary mRNA sequences.
What is the role of dicer in RNAi?
cuts dsRNA is cut into 21-25 bp fragments, acting as a molecular rule
What are two possible outcomes of siRNA or miRNA binding to RISC and then binding a complementary mRNA that would lower expression post transcriptionally?
Processed siRNA or miRNA, then associates with the RISC complex. One strand is degraded, and complementary mRNAs are destroyed, or complementary mRNAs are bound and prevented from being translated. This outcome is determined by how much sequence identity there is between the bound RNA and the mRNA.
Explain the RITS mechanism for how RNAi can alter transcription.
RITS complex carries the siRNA to the nucleus, where is can direct the complex to complementary nascent RNAs. When base pairing occurs, the complex recruits histone-modifying enzymes that close chromatin and spread heterochromatin. This is a mechanism that is extremely important for silencing centromeric repeats in S. pombe
What is the likely reason that RNAi evolved? What is one piece of evidence that supports this?
Current thinking is that RNAi evolved as a defense mechanism against the mutational effects of transposable elements. Mutations in the RNAi machinery can cause activation of normally silent transposable elements.
Contrast the approaches of forward genetics and reverse genetics. Which is more amenable to random mutagenesis? Which is more amenable to CRISPR-Cas9? Explain.
Forward genetics starts with a phenotype. You randomly mutagenize an organism, screen for individuals with an interesting phenotype, and then identify the gene responsible. This approach is highly amenable to random mutagenesis because it relies on unbiased mutations across the genome.
Reverse genetics starts with a gene. You deliberately disrupt, delete, or alter a known gene and examine the resulting phenotype to determine the gene’s function. This approach is highly amenable to CRISPR-Cas 9, which allows precise, targeted gene editing
Design a forward genetic screen for point mutations that cause cadmium resistance. Choose an organism, a mutagen, and a method to isolate both dominant and recessive mutations. Justify each of
your choices.
The organism could be yeast because it has a short generation time and can screen many individuals. I would mutagenize a large isogenic haploid yeast population with EMS to get point mutations. The mutagenized cells would then be plated onto cadmium-containing media so that only cadmium-resistant mutants survive. Dominant vs. recessive mutations can be distinguished by mating each resistant haploid to a wild type. If the diploid remains resistant, the mutation is dominant. If the resistance is lost, it is recessive. Resistant lines are then analyzed genetically and sequenced to identify the causal mutation.
Design a forward genetic screen for point mutations that cause increases in susceptibility to heat induced seizures. Choose an organism, a mutagen, and a method to isolate both dominant and
recessive mutations. Justify each of your choices.
The organism could be Drosophila because it reliably models temperature-sensitive phenotypes, can create lots of offspring, and has a short generation time. I would mutagenize an isogenic population with EMS because EMS mainly causes point mutations. F1 flies would be heat-shocked to identify dominant seizure-susceptibility mutations, since dominant alleles show their phenotype in heterozygotes. To isolate recessive mutations, each F1 line would be maintained over a balancer chromosome and intercrossed with carriers so that later generations produce homozygous mutants. These homozygotes can then be heat-shocked to reveal recessive seizure-susceptibility mutations. Finally, confirmed mutants can be mapped and sequenced to identify the causal mutation.
Why are F3 screens needed for identifying recessive mutations in organisms that cannot self, but these mutations can be identified in F2 screens in organisms that can self?
Recessive mutations are much more common, and identification is harder because they need to be homozygous. F2 screens are for organisms that can self (screen F2s for phenotype). F3 screen for organisms that cannot self (screen F2s for phenotype)
Explain the CLB strategy for finding recessive lethal mutations in Drosophila. Diagram out the three relevant crosses and indicate which genotypes survive and which die.
the CLB chromosome is a type of balancer chromosome. C= crossover suppression, L = recessive lethal, B = bar eyes mutation (dominant). The bar mutation permanently marks the CLB chromosome because recombination is suppressed.
Cross 1: mutagenized male and CLB female. Then isolate CLB females.
Cross 2: these females crossed to wild-type males. Different results for recessive lethal vs. all other mutations. If a lethal mutation, there will be no males. If it is not lethal, there will be a 2:1 female: male ratio.
Explain how loss of function in essential genes can be studied in haploids.
Conditional mutants allow for genetic screens in haploids to identify essential genes. Temperature-sensitive mutants allow for the study of essential genes under non-permissive conditions, thereby revealing their function when the protein is inactive. at room temp, there is no phenotype. At high temperatures, there is death. conditional loss of essential function, likely a missense mutation that slightly destabilizes an essential protein.
In a forward genetic screen for S. cerevisiae mutants that can grow in elevated concentrations of azole drugs, you identify a mutant that has moderate levels of resistance but also a severe growth defect in rich media. How would you go about identifying a secondary mutation that enhances the resistance phenotype? How would you go about identifying a mutation that rescued wild type growth in rich media?
to identify a secondary mutation that enhances the resistance phenotype, I would conduct a modifier screen by re-mutagenizing or evolving the mutant in high azole concentrations, then isolate and sequence resistant mutants.
For the suppressor mutation that restores growth in rich media, I would select for fast-growing colonies in rich conditions, sequence these suppressors, and identify the mutation through co-segregation with the restored growth.
Describe the two main mechanisms of synthetic lethality. Can three genes display synthetic lethality? Explain
Genetic redundancy/parallel complementary pathways are when several genes perform redundant functions. Loss of function of one can be compensated for by the other (and vice versa). Loss of function of both genes leads to loss of essential function and death. Null alleles can display this property.
Within pathway interactions are when several genes perform sequential actions in an essential pathway. Loss of function in one component leads to reduced activity but not lethality. Loss of function in both components leads to greatly reduced activity and is lethal. Null alleles cannot display this property, and the null allele would block the pathway by itself and be lethal.
Yes, three genes can display synthetic lethality. In cases where two genes provide parallel functions and a third gene disrupts this redundancy, the combination can lead to lethality.
Explain how sequencing can be used to identify causal mutations in a forward genetic screen
Sequencing a large cohort of F2s harboring a particular mutation/phenotype can lead to the identification of the causal mutation. All affected individuals should share the causal mutation. Recombination will lead to variable presence of all other induced mutations.
What two problems with forward genetic screens does experimental evolution circumvent?
The biased mutation spectrum of the chosen mutagen and the ecological relevance of mutants are often questionable. Experimental evolution circumvents both of these issues. The organism in the lab with deliberate selective pressures by the natural spectrum of mutations is used, and costly mutations will often be selected against
What is the function of Cas9? tracrRNA? crRNA?
Cas9 induces double-stranded breaks at specific sequences. Cas9 allows for these site-directed breaks via complexing with a guide RNA.
The CRISPR-Cas system is a defense mechanism against invading nucleic acids. Spacers are sequences derived from phage. These spacer sequences are used by the Cas protein as guides.
CRISPR sequences are transcribed into noncoding RNA. These are then processed into crRNAs that consist of the unique spacer sequence and the identical repeat steps. Processing is done by cas encoded RNAases.
Upstream of the CRISPR locus is another gene that is transcribed into tractRNA. tractRNA is partially complementary to the repeat sequences and forms a stem loop that binds Cas endonuclease. tracrRNAs facilitate crRNAs binding to Cas.
If a bacterium survives a phage infection, what is a likely outcome for its CRISPR array? Is this change heritable?
The bacterium is likely to incorporate a new spacer sequence derived from the phage into its CRISPR array, providing immunity against future infections. This change is heritable, as the modified CRISPR array will be passed down to its progeny.
You have a stock of wild type C. elegans. Suppose you want to assay the effects of loss of function of a particular gene in this stock. Propose an experiment to accomplish this without altering the genotype of your wild type stock.
You can perform a RNA interference (RNAi) experiment where you introduce double-stranded RNA corresponding to the gene of interest. This will induce degradation of the target mRNA, effectively resulting in a loss of function for that gene without altering the genotype of the wild type C. elegans.
You are studying a gene in mice that is important for the proper formation of the brain, and this gene is only present in the brain in the wild type. You construct two chimeric alleles, one with the gene under the control of a promoter that causes this gene to be expressed throughout the body later in development, and another that causes high expression in most areas of the early embryo. The first gene yields viable mice, but upon inspection, you find that the concentration of a cell type called astrocytes in the spinal cord has increased dramatically. The second allele leads to early embryonic developmental arrest and death. What, if anything, can you infer from each of these alleles?
The first allele suggests that the gene's expression in non-brain tissues, such as the spinal cord, can lead to abnormal increases in specific cell types like astrocytes, indicating a regulatory role beyond normal brain function. The second allele indicates that proper timing and location of gene expression are critical for early development, and misexpression can lead to lethal consequences.
Why compared to many other areas of medicine, does medical genetics need to have a strong emphasis on the patient’s family in addition to the patient?
because the family’s role in care is critical for many genetic diseases, and risk needs to be assessed and communicated to the family regarding future children in the immediate and extended family
Design an assay that would allow for the disease allele of the huntingtin gene to be identified using PCR.
Design PCR primers that flank the CAG repeat region in exon 1 of the HTT gene. When you run the PCR product on a gel, the disease allele shows a larger fragment because it contains an expanded CAG repeat. Comparing fragment sizes allows you to identify whether a person carries the Huntington disease allele.
Why does presymptomatic testing of Huntington’s disease entail complex ethical issues?
Presymptomatic testing is determining the genotype for diseases with a late stage of onset. Huntingtons disease presents many unique challenges and questions. wihtout the information, it may be more difficult to make future plans if the disease progresses. wiht the information, learning you have the disease allele will be life altering and traumatic.
Presymptomatic testing gives people the option to choose, but it can also raise ethical issues for their family.
What is maternal serum sampling? Is it a diagnostic test? Why nor why not?
maternal serum sampling is measuring the levels of three proteins in the mother’s blood. Drawing blood is all that is required. Serum sampling is a screening test, not a diagnostic test (false negatives and false positives occur).
Why is fetal cell sorting in principle easier for male fetuses? Describe two reasons why fetal cell sorting is very difficult.
Fetal cell sorting is easier for male fetuses because male fetuses contain the Y chromosome, which makes them distinct from the mother’s chromosomes.
Two reasons fetal cell sorting is very difficult are the low concentration of fetal cells in maternal blood and the challenge of differentiating between fetal and maternal cells due to their similarities.
What are amniocentesis and chorionic villus sampling? Would these procedures typically be recommended before an abnormal result from a noninvasive test? Why or why not?
Amniocentesis is when a needle is used to pulls amniotic fluid from the mother’s uterus. The fluid contains fetal-derived cells that can be cultured and subjected to chromosome analysis. With this, there is a risk of spontaneous abortion or fetal injury.
Fot chorionic virus sampling, a small tube is passed transvaginally to extract cells from the chorion (part of the cell that is fetal-derived). There is also a risk for spontaneous abortion or fetal injury.
Describe the process of preimplantation screening and how it can be used to mitigate the risk of a child with a genetic disease. In what context is this method typically used?
in vitro fertilization (IVF) is done by injecting a female with hormones to induce ovulation. Eggs from the surface are then removed and either frozen. The eggs will undergo several cell divisions over 3-5 days and then be implanted into the uterus. This is less than 50% effective and is very expensive per session.
This can be used to mitigate genetic disorders because if both parents are carriers for a genetic disease, a single cell can be removed at the 8-16 cell stage, and then the genotype of the embryo can be determined. Those with the genotype would be excluded because they would develop the genetic disease
What is the Guthrie test? How does it work?
a test to screen for PKU. It is a test that harvests a few drops of newborn blood via a heel stick. The blood is plated on a bacterial culture with a toxic phenylalanine analog that can be misincorporated into proteins. PKU-affected individuals have abnormally high levels of phenylalanine in the blood. drops of their blood restore bacterial growth.
Why are population carrier screens performed for Tay-Sachs disease? What is the molecular basis of pathogenesis in Tay-Sachs disease?
This is done to identify phenotypically normal individuals who are carriers for a disease because some subpopulations of humans have higher frequencies of diseased alleles. For example, Ashkenazi Jewish populations have a high frequency of Tay-Sachs disease.
Tay-Sachs is a recessive lethal neuromuscular disorder that causes death in early childhood. This is caused by a loss of functional hexosaminidase, leading to a buildup of GM2 ganglioside and killing nerve cells. This causes infants to lose the ability to turn over, crawl, hearing loss and Living past 3-5 years is extremely rare.
Are direct-to-consumer genetic tests diagnostic? Why or why not?
Direct-to-consumer genetic tests are not considered diagnostic because they typically provide risk assessments rather than definitive diagnoses. These tests can indicate potential genetic predispositions but do not confirm the presence of a genetic condition without further clinical evaluation.
Describe one potential drawback of direct-to-consumer genetic testing in the absence of the support of a genetic counselor.
Without the guidance of a genetic counselor, individuals may misinterpret their genetic information, leading to unnecessary anxiety or false reassurance. They may also lack access to reliable follow-up options for testing and care.
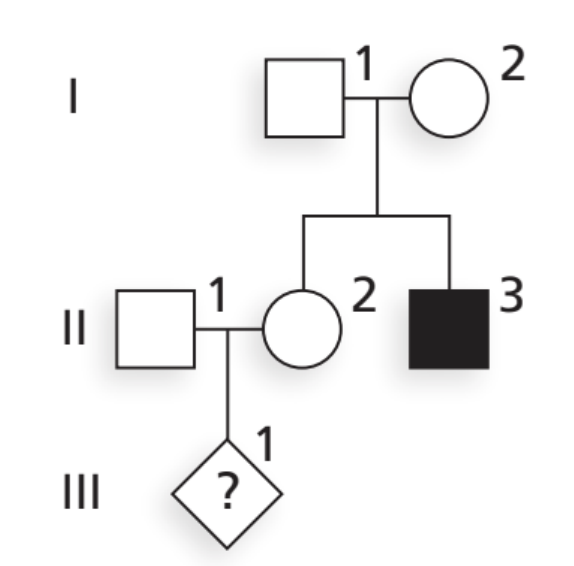
Given the following pedigree concerning an autosomal recessive condition with a carrier frequency of 3.2%, calculate the probability that individual III-1 will be affected by the disease. How does this compare to the rate of disease expected for a random mating? Formulate a 2-3 sentence explanation of these odds as if you were a genetic counselor and II-1 and II-2 were in your office seeking guidance.
Based on your family history, your chance of having a child affected with this condition is about 1 in 190, which is higher than the general population risk of about 1 in 3,900. This increase is because one of you has a higher chance of being a carrier due to having an affected sibling. If you’d like more precise information, we can arrange carrier testing for both of you, which would let us refine this risk estimate further.
P(III-1 affected)=P(II-2 carrier)×P(II-1 carrier)×41=32×0.032×41≈0.0053
How is cancer a “disease of the genome”?
cancer is uncontrolled cell growth leading to tissue dysfunction.
Cancer can be thought of as natural selection playing out among somatic cells. Explain this idea.
Cancer develops when mutations give certain cells a survival advantage, allowing them to proliferate unchecked. This process resembles natural selection, where the most fit cells dominate, leading to a more aggressive tumor.
Define proto-oncogene, tumor suppressor gene, angiogenesis, metastasis, driver mutation, and passenger mutation
A proto-oncogene is a gene that promotes cell cycle progression (gas pedal)
A tumor suppressor gene is are genes that normally act to slow/stop cell cycle progression, “brakes”.
Angiogenesis induction is the recruitment of normal cells to form blood vessels that supply the tumor with oxygen
Metastases are cells that invade tissues other than the one the tumor began
driver mutations are mutations that give rise to a cancerous phenotype
passenger mutations are mutations that are present in the tumor but do not drive cancer progression.
Why is age the biggest risk factor for cancer?
Every cell division introduces a small chance of error. As you age, the more your cells divide, giving them more opportunities to have division errors. Most cancers require multiple genetic “hits,” which simply take time to occur. Aging tissues also experience chronic inflammation and epigenetic changes that further promote the development of cancer.
Contrast sporadic and familial cancers. How do they differ?
Sporadic cancers are caused by many mutations that progressively transition normal cells into cancerous ones. Familial cancer is where the presence of a germline mutation predisposes individuals to a higher risk of a certain cancer.
Explain the cause of chronic myelogenous leukemia. How can it be treated by Gleevec?
Chronic myelogenous leukemia is caused by a reciprocal translocation between chromosomes 9 and 22. This genetic alteration results in the BCR-ABL fusion protein, which promotes cell proliferation and inhibits apoptosis. Gleevec (imatinib) treats this leukemia by targeting and inhibiting this protein, effectively controlling cancer cell growth.
Explain Knudson’s two hit hypothesis using the example of retinoblastoma.
Retinoblastoma is a rare cancer of the retina that affects 1/100,000 newborns and young children. Knudson’s two-hit hypothesis posits that for cancers like retinoblastoma to develop, both copies of the RB1 gene must be mutated. The first ‘hit’ can be a hereditary mutation present at birth, while the second ‘hit’ often occurs later due to environmental factors or additional genetic changes.
Li Fraumeni syndrome can lead to inheritance of susceptibility to many types of cancer. What explains this?
Li fraumeni syndrome is an inherited susceptibility to many types of cancer. This is caused by mutations in TP53, which encodes p53, a transcription factor. This is a tumor suppressor gene, leading to a two-hit problem. Only a single mutation in TP53 is sufficient to drive cancer development in a heterozygote. p53 acts to ensure that cells with DNA damage do not divide. It causes G1 arrest and can lead to apoptosis if the damage is not repaired. Without functional p53, cells with damage proliferate and accumulate more mutations, leading to cancer.
Most cases of breast cancer are sporadic. What can indicate that a case is due to inherited susceptibility rather than being sporadic?
Bilateral cancer or cancer at a young age may signal inherited susceptibility. Contralateral breast cancer, which is cancer in the second breast subsequent to cancer development in the first breast, may also signal inherited susceptibility.
Why do mutations in BRCA1 and BRCA2 lead to increased risk of cancer? Is this similar to why TP53 mutations lead to increased risk of cancer? Explain.
BRACA1 and BRACA2 mutations influence inherited susceptibility. LOF mutations in these genes increase susceptibility because they increase the mutation rate, making that cell lineage more prone to subsequent cancer-causing mutations. Neither mutation guarantees the development of cancer, other genes and environmental factors are at play
Why are certain cancers not treatable by surgery? Why do chemotherapeutics typically target rapidly dividing cells?
some cancers are not easily removable due to location of gene and type of tumor, Chemotherapeutics target rapidly dividing cells because such cells are more susceptible to damage during the cell cycle, which is where many chemotherapy drugs exert their effects.
Explain the logic of CAR-T cell therapy to treat ALL. Explain the dual consequences of on-target off tumor cytotoxicity of these cells against CD-19 positive B-cells.
CAR-T cells are engineered to recognize and destroy cancer cells expressing the CD19 antigen. While effective against acute lymphoblastic leukemia (ALL), they can also harm normal B-cells that share this antigen, leading to potential immune deficiencies.
What is a restriction enzyme? What are the normal functions of restriction enzymes in bacteria? Why do they not cut the host chromosome?
A restriction enzyme is an enzyme that makes double-stranded cuts in DNA at specific sequences. They were discovered in bacteria and serve to protect them from invading nucleic acids. These enzymes restrict the growth of bacteriophages.
Bacteria protect their own DNA by adding methyl groups to it. Restriction enzymes only cut DNA that is not methylated, so they destroy invading DNA (like viruses) but leave the bacteria’s own/ “self” DNA alone.
Define vector in the context of recombinant DNA technology.
A vector is a carrier molecule of DNA with attributes that allow it to be replicated in a biological system. The vector is inserted with the DNA of interest into a living organism that amplifies the DNA as it replicates.
Why is it true that the use of a single restriction enzyme on DNA from two sources will yield multiple possible ligation products? Why is this not going to happen in directional cloning?
Using a single restriction enzyme gives all fragments identical sticky ends, allowing them to ligate in many possible combinations and orientations. In directional cloning, two different enzymes create two different sticky ends on both the vector and the insert, so fragments cannot re-ligate incorrectly. Ligation can occur in only one orientation and self-ligation is prevented.
Why is directional cloning a much better option for cloning into expression vectors as compared to using a single restriction enzyme?
Directional cloning is better than using one restriction enzyme because directional cloning creates non-identical, non-compatible sticky ends, so the insert can ligate into the vector in only one orientation. In contrast, using a single restriction enzyme makes identical complementary ends, allowing the insert to ligate in either orientation and generating unwanted or nonfunctional ligation products.
You are cloning a human gene responsible for sequestration of heavy metals in the cytoplasm. You use a pUC-based plasmid. Following construction of your recombinant vector, your transformation only yielded blue colonies. What does this indicate?
Blue colonies indicate that the lacZ α-fragment remained intact, meaning the insert did not disrupt the MCS. This occurs when the vector re-ligates without the insert, so blue colonies signal that the cloning was unsuccessful.
What is the key difference between a plasmid and a BAC?
BAC is a bacterial artificial chromosome that is larger than a plasmid and can insert ~100-200 kb. Plasmids are typically smaller, allowing for inserts of ~1-10 kb. BACs accommodate much larger DNA fragments, making them useful for cloning larger genes or genomic DNA.
Suppose you sequence all clones in a gDNA and a cDNA library derived from the same organism. In the gDNA library, you find sequences corresponding to gene A are three times as abundant as sequences corresponding to gene B. However, in the cDNA library there are three times as many sequences corresponding to gene B compared to gene A. Propose a plausible explanation for this pattern.
Gene A appears often in the gDNA library because it has more copies in the genome. It could be a part of a gene family, recently duplicated, or present in multiple tandem repeats.
Gene B appears more often in the cDNA library because it is more highly expressed, producing more mRNA that is converted into cDNA.
These reflect gene copy number in gDNA vs. expression level in cDNA, which are independent features of the genome.
You are trying to express a human gene in bacteria. You find that no matter how strongly you induce expression of the human gene, there is very little protein produced and some of your data seem to indicate that the protein is not folding properly in its bacterial host. This is in stark contrast to the amount of mRNA for this gene being produced, which is very high. Propose a plausible reason for
this pattern and devise a solution
The most likely explanation is poor translation or misfolding of the human protein in bacteria. This is often due to rare codons or improper folding. The solution is to codon-optimize the gene or adjust expression conditions (such as lowering induction temperature) to improve folding and yield.
Suppose the genetic code were not universal. What complication would this introduce in terms of recombinant DNA technology? Would it make heterologous gene expression impossible? Explain.
If the genetic code were not universal, it could lead to issues with translating heterologous genes in different host organisms. This would mean that genes from one organism might not be accurately translated or expressed in another due to differences in codon usage, potentially affecting protein production and functionality. While it might complicate heterologous gene expression, it would not make it completely impossible; adaptations or modifications could still be made.