DNA Replication BSCI222
1/58
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
59 Terms
Termination
process of elongation ends when template strand is no longer single stranded
how does termination occur in prokaryotes?
Meets another Okazaki Fragment or meets the origin on the leading strand
how does termination occur in eukaryotes?
meets the end of the chromosome
how many or origins of replication (ori) in prokaryotes
one
how many of origins of replication (ori) in eukaryotes?
multiple per chromsome
replicon
segment of DNA replicated one origin per replicon
when does replication terminate (per replicon)?
when bubbles of different replication forks meet
what do replication licensing factors do?
license origin sequences during G1 allowing them to function as origins of replications
- license is lost after replication begins
do eukaryotes and prokaryotes have the same enzymes for replication?
no, eukaryotes have different enzymes (do not need to know for this class)
why does telomere shortening occur?
primer is removed but there is no longer a 3' end for DNA Poly III to add to on lagging strand
what does telomere shortening do?
overtime leads to mutations or removal of euchromatin
- possible cause of aging and may play a role in cancer development
STRs
short tandem repeats (found in telomeres)
telomerase
has RNA and proteins which can serve as a template for extension of 3' end of telomere so DNA Poly III can add nucleotides on the lagging strand
- ONLY ACTIVE IN PROLIFERATIVE CELLS (fetal cells, bone marrow, germ line, intestinal)
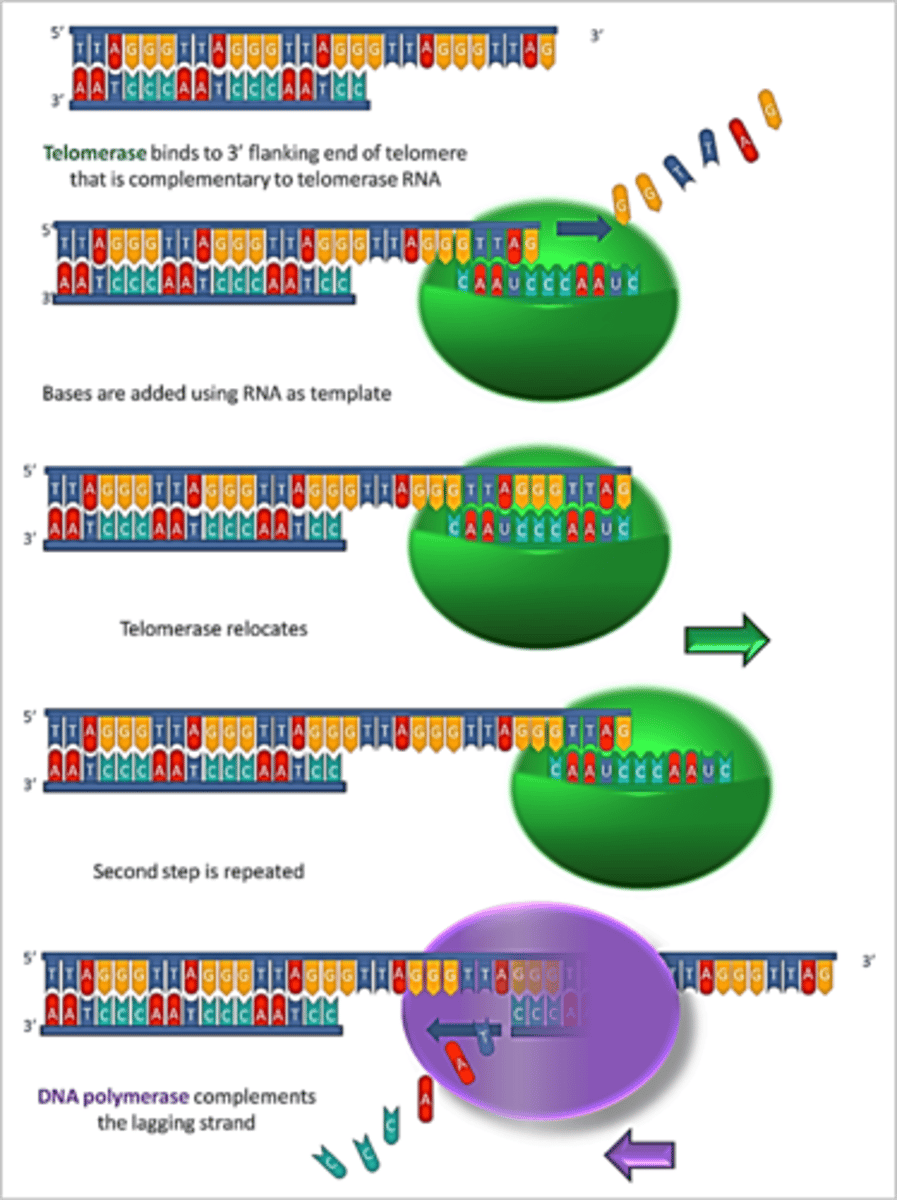
what is the nucleotide common telomere STRs for vertebrates?
TTAGGG
what is PCR?
polymerase chain reaction makes a ton of copies of same DNA segment
how does PCR work?
DNA is heated in a solution of dNTPs, DNA Poly (Taq), primers to replicate DNA
DNA Polymerase (Taq)
used in PCR since it can operate at a higher temperature
what do primers do in PCR?
add to a ssDNA to form a primer for DNA Poly (Taq) to add nucleotides to, must be antiparallel to template
- only needs to be ~22 bp long to find a unique position in human genome
steps for PCR
1. heat DNA ~90C to break H-Bonds
2. Cool to ~60C for primer to add
3. Heat to ~72C for DNA Poly (Taq) to work
4. Repeated 30-50 times to create millions of replicas
what is gel electrophoresis used for?
separation of DNA fragments by size
electrophoresis
carry with electricity
- negative charged DNA moves towards positive electrode
how to compare band location on gel electrophoresis?
use a ladder with known bp sizes
how can PCR be used to tell apart STRs?
homologous chromosomes will have same motif (repeat) but in very different lengths
- example: crime scene gets STR DNA and can compare it against suspects since lengths of motifs will be the same
motif
repeated segment of DNA
locus (loci plural)
location on a chromosome
- location is the same for homologous chromosomes
minisatellite repeats
motifs 5-10bp long
microsatellite repeats
motifs 2-4bp long
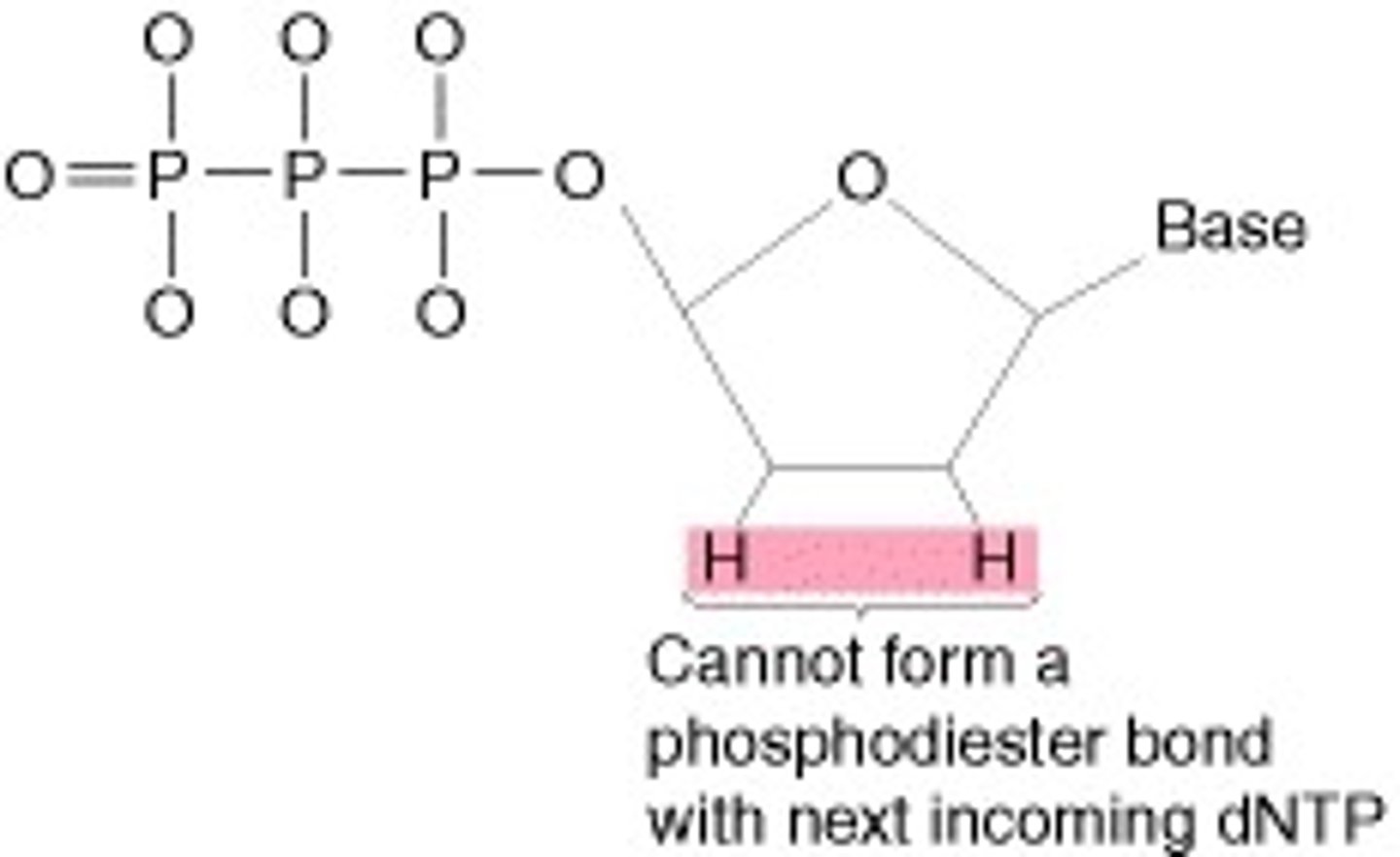
Dideoxy Nucleotides
chemically modified nucleotides that halt the DNA replication process to determine the sequence of an unknown piece of DNA
- lack a 3' hydroxyl
- dideoxy = lacking 2 hydroxyls
Dideoxy/Sanger Sequencing
can be used to sequence a gene
How does Dideoxy/Sanger Sequencing work?
uses many normal dNTPs and ddNTPs to replicate a sequence, enzymes pick at random which to use giving a variety of lengths to sequences
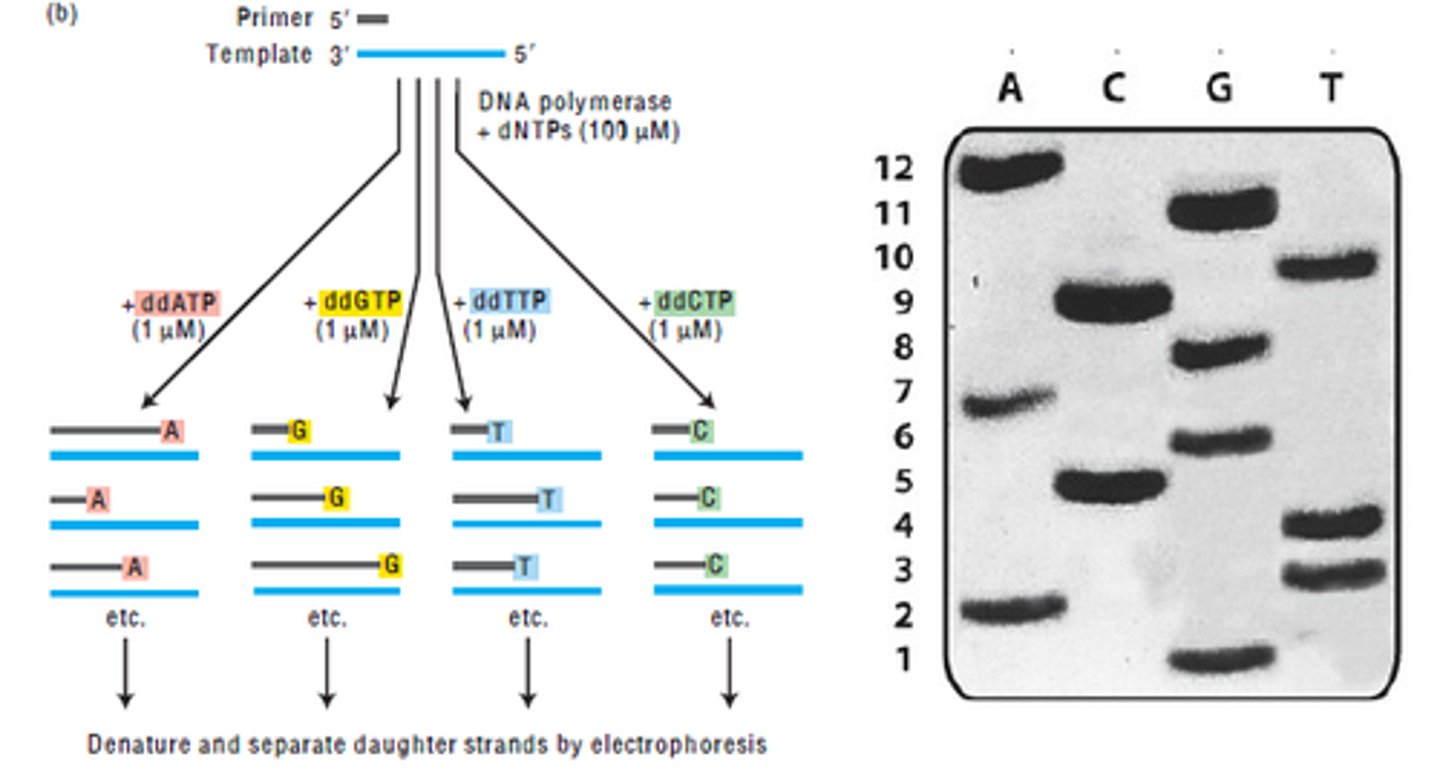
how to analyze Dideoxy/Sanger sequences?
tagging ddNTPs with florescent dyes or gel electrophoresis
Illumina Sequencing
Use dNTPs with base specific dyes and reversible terminator
how does Illumina Sequencing work?
1. fragment of DNA + universal primers to prime a variety of locations
2. add all dNTPs, one dNTP will add to the synthesis strand and will glow, replication will stop bc of terminator
3. both the fluorescent and terminator are cleaved off of the nucleotide
4. repeat until sequence is completed
dna replication is a ______ process
semi- conservative
- template strands are kept intact throughout the process
what are the 4 stages of DNA replication
1. initiation
2. unwinding
3. elongation
4. termination
initiation
regulates process of replication; creates space for enzymes to attach
unwinding
separation of the dna strands, allowing synthesis proteins access
elongation
synthesizes new dna strand through complementary bp addition
termination
when template becomes fully double stranded, polymerases are blocked
what proteins initiate replication and what do they do?
Initiator proteins (dnaA in bacteria) bind at ori creating an opening
helicase
An enzyme that untwists the double helix of DNA at the replication forks and is powered by ATP
SSBPs
bind to separated bps to ensure they do not recombine
DNA gyrase
a type of topoisomerase that is found in bacteria
- relieves positive supercoiling ahead of the replication fork/in front of helicase
triphosphate nucleotides
Building blocks for RNA and DNA synthesis
DNA primase
places RNA temporary placeholders allowing DNA Poly III to bind and start elongation
- place holders are complementary to replication fork
DNA Polymerase III
adds nucleotides to the new strand in the 5'->3' direction and fixes mistakes during elongation
DNA Polymerase I
replaces RNA Primers with DNA and fixes mistakes after elongation of a segment
Ligase
binds newly added DNA from DNA Poly I to the rest of the strand
mechanism for -TP forming a phosphodiester bond
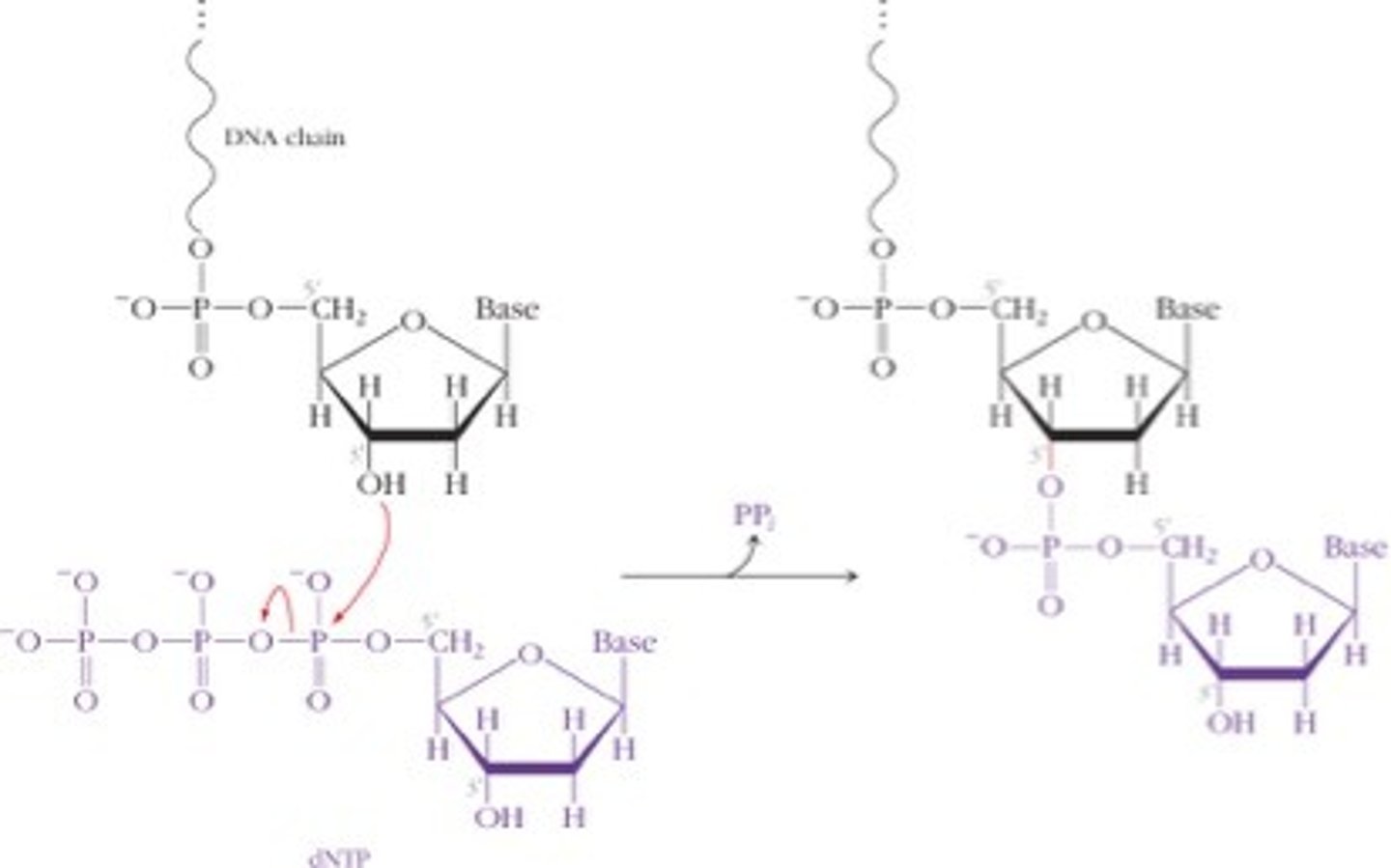
phosphodiester bond
the type of bond that links the nucleotides in DNA or RNA. joins the phosphate group of one nucleotide to the hydroxyl group on the sugar of another nucleotide
what are the limitations of DNA synthesis?
1. need an existing 3' end to start
2. direction is ALWAYS 5' -> 3'
which enzymes can work as an exonuclease in the 5'->3' direction?
ONLY DNA Poly I
which direction does DNA Poly I work in polymerizing nucleotides?
5' -> 3'
what happens of DNA Poly III detects something wrong in a placed nucleotide?
can enter "editing" mode and work 3' -> 5' (BACKWARDS) to function as an exonuclease
exonuclease
enzyme that cleaves off nucleotides
which enzymes can act as an exonuclease in the 3' -> 5' direction?
BOTH DNA Poly I and DNA Poly III
leading strand
synthesis (5'->3') in the same direction as replication fork, continuous
- one rna primer
lagging strand
synthesis (5'-3') in the opposite direction as the replication fork, not continuous
- many rna primers
okazaki fragments
Small fragments of DNA produced on the lagging strand during DNA replication
- each require its own primer