BMS1062 - W7: Regulation of Gene Expression
1/22
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
23 Terms
Why are not all genes expressed at the same time?
Prokaryotes:
different genes expressed in response to different environments
presence of a food substance (e.g. lactose)
don’t waste energy expressing genes which are of no use in a particular environment
Eukaryotes:
All cells retain the entire hereditary potential (exactly the same set of genes in it) of the organism, they differentiate into different types of cells (e.g. brain vs muscle) so need to express different factors.
How is gene expression controlled in prokaryotes vs Eukaryotes?
Prokaryotes: Ground state = on
The normal state of a prokaryotic gene is called the on state.
prokaryotic genes always ready to be expressed all the time
just needs RNA polymerase to bind and start transcription - RNA polymerase is usually all that is required to get gene expression going
Eukaryotes: Ground state = off
Default state of gene expression in eukaryotes is off state.
generally not ready to be transcribed in eukaryotes
need RNA polymerase, but also almost always need a large number of other proteins that act as transcription factors and enhancers
all these additional proteins are required for the eukaryotic RNA polymerase to bind efficiently to promotor and get into an active form so it can start transcription
we also say that the ground state of eukaryotic gene is off because eukaryotic DNA in nucleus is very tightly compacted and not easily accessible to RNA polymerase (DNA form chromatin, then nucleosomes, then chromosomes)
So only a small fraction of eukaryotic genes is expressed at any point in time for 2 reasons
most of the DNA is tightly condensed and not accessible to RNA polymerase
activation of RNA polymerase for transcription requires a whole bunch of transcription factors and enhancers
Considering the eukaryotic DNA is so compact, how do eukaryotic genes become accessible to transcription proteins?
Achieved through local relaxation of some portions of the chromatin by ‘chromatin remodeling proteins’
Prokaryotic gene expression:
Bacteria can change their gene expression in response to environment very rapidly.
e.g. metabolism of different sugars requires different enzymes - encoded by different genes.
must be able to sense when sugar is present, up-regulate necessary genes in response, downregulate necessary genes when sugar is absent
Different genes can be regulated in different ways but share some common mechanisms
Regulation at level of transcription
most common form of regulation
transcription is DNA → mRNA
can regulate how much mRNA is made from a gene (no mRNA = protein not expressed)
Regulation at the level of translation
Regulation at the level of transcription:
Requires a promotor sequence (promotor sequences determine where RNA polymerase starts transcription
e.g. TTGACA-17bp-TATAAT (most common and likely one of the strongest promotors)
promotor upstream of gene helps determine how efficiently RNA polymerase binds there and how much gene product is made
RNA polymerase is recruited to the promoter by the sigma factor (sigma factor recognizes and binds to promotor, then it recruits RNA polymerase and binds to both sigma factor and DNA)
different types of sigma factor (but only one RNA polymerase)
depending on the exact nucleotide sequence of promoter, different promoters bind different sigma factors.
e.g. sigma factor 32 binds to genes induced by heat shock. When temp rises, levels of sigma factor 32 rapidly rise and promoters of all genes encoding heat shock response process will be bound to sigma factor 32 and they will all get transcribed. Because there is limited amount of RNA polymerase molecules in the cell, its use for high temp conditions will be biased towards heat shock proteins.
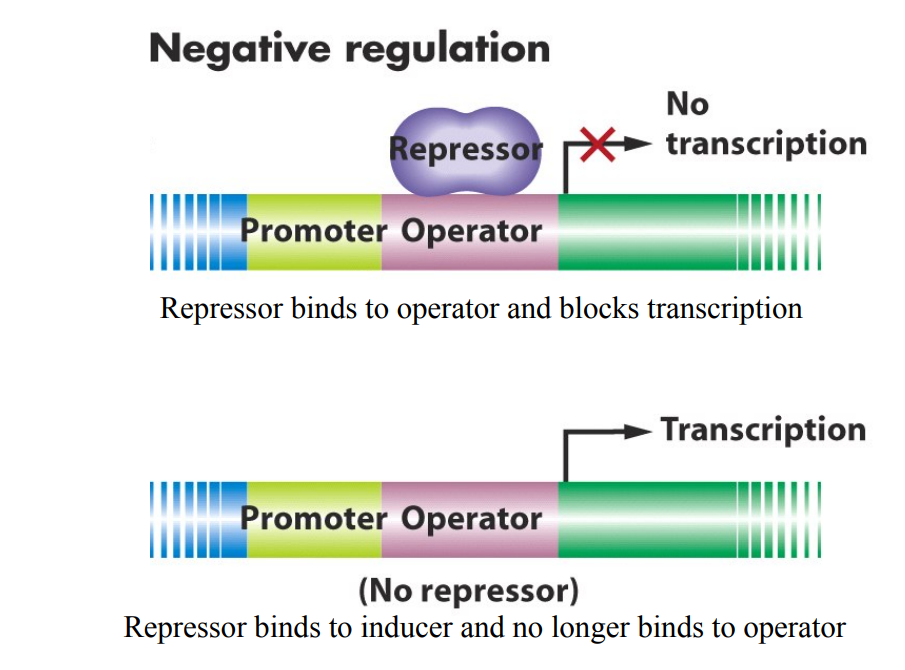
Negative regulation: Repressor
carried out by a repressor protein
transcription is stopped or reduced when repressor protein is bound (turns promotor OFF)
when genes need to be turned on, negative regulator must be inactivated
Positive regulation: Activator
is required before the gene is efficiently transcribed
activator is required to bind to the DNA to help RNA polymerase bind (to turn the promoter on)
Negative regulation (Repressors):
What is an operator site?
What is an inducer molecule?
Repressor protein binds to DNA turning promoter off (RNA cannot bind and transcription cannot occur)
Operator site: The place where the repressor binds
operator sequence often partially overlaps promotor sequence which stops RNA polymerase from binding to promotor
Repressor may bind an ‘inducer’ molecule (sometimes called effector?)
binding of inducer to repressor inactivates the repressor (repressor falls off as it cannot bind to the DNA)
Transcription can occur
Co-repressor:
in some cases, co-repressor if required to allow repressor binding
transcription cannot occur
e.g. LacI (repressor protein)
binds between -5 and +21 (reminder that transcription starts at +1)
because the LacI binds just before the start of transcription:
the RNA polymerase cannot bind very efficiently
even if it can bind, it cannot unwind the DNA to start transcription
Lac repressor protein can bind an inducer called allo-lactose
when the inducer binds to LacI, it changes shape of LacI and can no longer bind to operator site.
Now RNA polymerase can bind and transcription can occur
Positive regulation: Activator
Activator protein is required to bind the DNA to turn promoter on
promoter only active when activator binds
activator often can only bind to DNA in the presence of the inducer molecule (inducer interacts with activator helping it bind to the DNA)
in absence of inducer, gene will be OFF
activator binds usually upstream of the promoter sequence (can interact with RNA polymerase and help it bind to DNA)
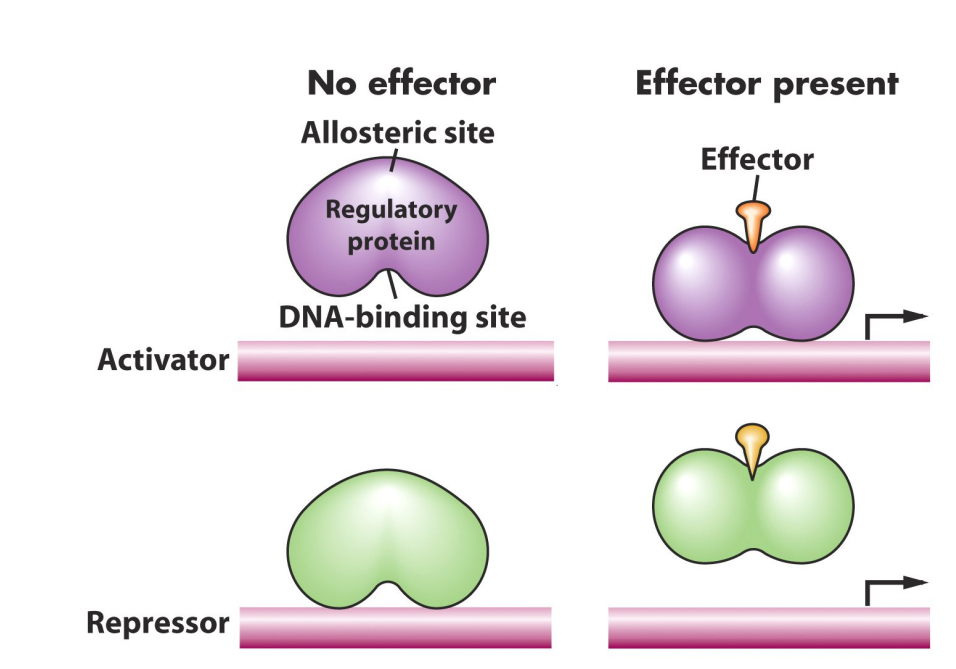
Allosteric proteins:
Allosteric proteins are proteins that exist in two different conformational forms (different shapes)
transcriptional repressors and activators are allosteric proteins.
These allosteric proteins have at least two different binding sites
one for binding inducer
one for binding DNA
These proteins change conformation when bind to inducer (changing ability to bind DNA)
What is an operon?
A set of genes that are transcribed together on the same transcript
Lac operon:
Controlled by both positive and negative regulation.
Lac operon of E. coli controls metabolism of lactose (a disaccharide made of glucose and galactose units)
takes a bit more energy as galactose needs to be changed to a metabolizable sugar
Negative regulation:
lac repressor (product of lacI gene)
Inducers are low MW beta galactosidase (allo-lactose)
Positive regulation:
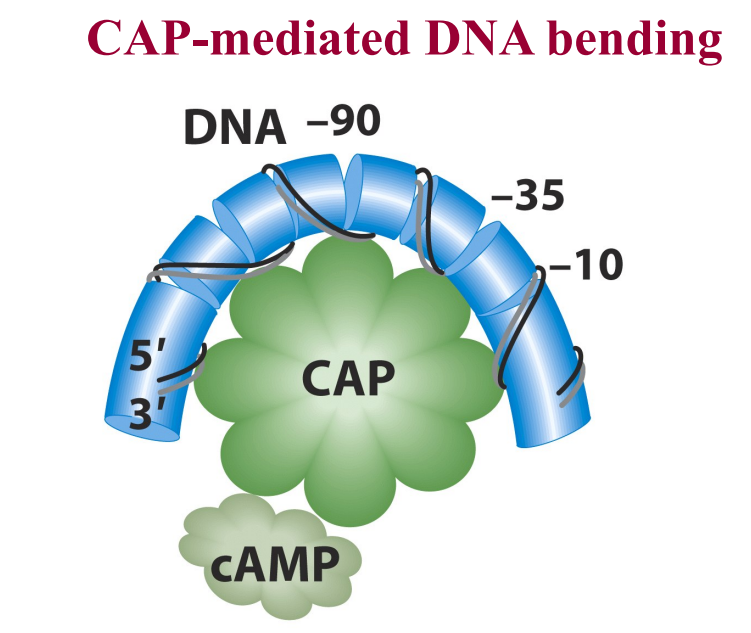
by CAP protein (catabolite activator proteins)
inducer is cAMP (which responds to glucose - sense how much glucose)
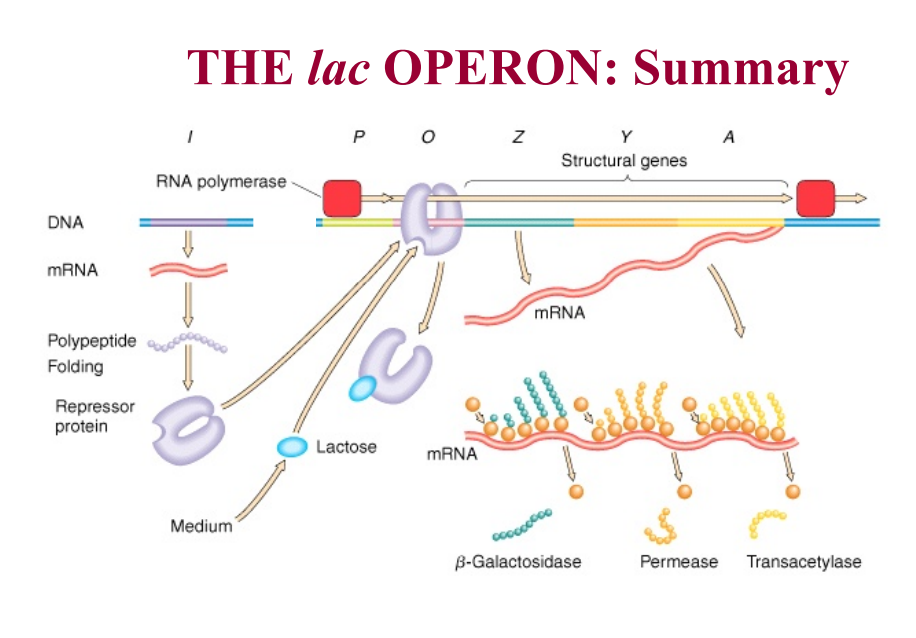
Lac operon has 3 genes that are all under control of one promoter
the three genes are all associated with the metabolism of lactose, so you want to turn them on/off together.
always turn on/off together
only see operons in prokaryotic systems
The three genes:
LacZ: encodes for beta galactosidase (hydrolyzes lactose to glucose and galactose)
Lac Y: encodes lactose permease (transports lactose into the cell)
LacA: encodes galactoside transacetylase (biological role is unknown)
Negative regulation of Lac operon:
LacI: encodes Lac repressor
repressor protein made from the LacI gene which is found elsewhere in chromosome
made all the time
in absence of lactose, repressor is active, binds to operator, and blocks unwinding of DNA
when lactose is present, it is converted to allo-lactose and binds to allosteric site on Lac repressor - changes its conformation and it can no longer bind to Lac operator site - RNA polymerase is now able to transcribe the genes
Positive regulation of Lac operon
glucose is preferentially metabolized over lactose
lac operon will not be expressed when glucose is present
Mediated by levels of cAMP and positive regulator CAP
cAMP (cyclic AMP)
Glucose levels regulate cAMP levels
high glucose inactivates adenylate cyclase so ATP is not converted to cAMP (high glucose = low cAMP)
low glucose No inactivation so ATP → cAMP (low glucose = high cAMP)
cAMP is inducer of CAP activator protein
CAP needs cAMP to bind
CAP:
Cap is an allosteric protein
activates transcription after binding cAMP
induces DNA bending which allows DNA polymerase to access promoter more efficiently and partially unwind
CAP only activates transcription efficiently in the absence of Lac repressor
So for high LAC operon expression need:
glucose absent
lactose present
Under the following conditions: What occurs:
a) Glucose present, no lactose
b) Glucose present, lactose present
c) glucose absent, lactose present
a) No expression of lac mRNA
repressor can bind as lactose is absent (which is the inducer which prevents repressor from binding)
CAP does not bind as glucose = low cAMP which is inducer for activator
b) little expression of lac mRNA
glucose = high, cAMP = low, meaning inducer is not binding to activator, meaning gene expression is not occurring
lactose present, repressor become inactivated, repressor is not bound (which typically would allow for gene expression) but CAP will not bind. So only little bit of expression
c) gene expression
cAMP levels are high, binding to activator, binding to DNA and activating transcription.
lactose present, repressor become inactivated, repressor is not bound = gene expression
Lecture 14:
Why are some genes inducible and some repressible?
This is to do with whether they are responsible for breaking molecules up or joining them together. (destructive or constructive side of metabolism)
Destructive side - catabolism (break down)
e.g. Lac operon (inducible gene)
when substrate is present, the substrate is an inducer (induces gene expression)
Constructive side - anabolism (biosynthesis)
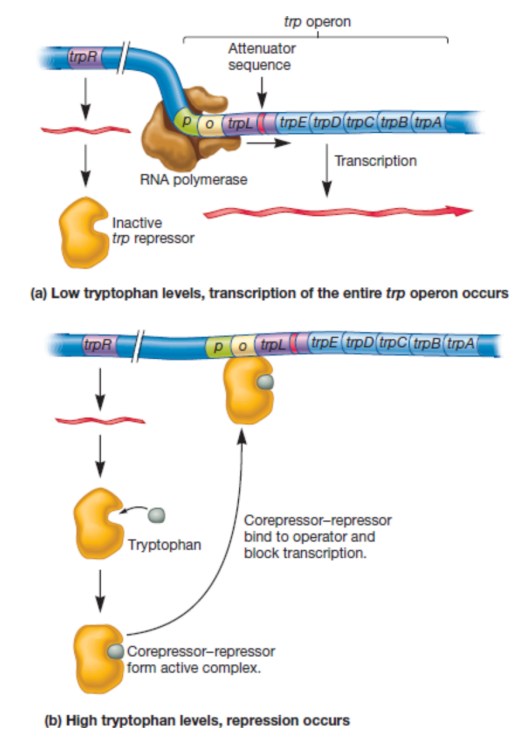
e.g. Trp operon (repressible gene)
when end product is present, the product (tryptophan) is a co-repressor (because you already have so much of it, why make more)
What is feedback repression?
What is feedback inhibition?
Feedback repression: The negative control of gene expression.
related to gene expression
how anabolic (biosynthesis) pathways are regulated
where the product of a pathway interacts with a regulatory protein (e.g. a repressor) to stop transcription of genes encoding enzymes in the pathway)
Can be achieved through two mechanisms:
regulating transcription initiation
regulating transcription termination
Feedback inhibition:
related to protein activity
where the product of a pathway inhibits the activity of an enzyme earlier in the pathway
often end product of the pathway will bind to an enzyme for the first step reducing its activity
Tryptophan Biosynthetic pathway in E. coli
multistep pathway
all enzymes are encoded by genes that belong to a single operon (with one promoter)
Feedback inhibition:
The end product (Tryptophan) binds to the allosteric site of the first enzyme trpE and inhibits its activity.
All the genes are still being expressed to make each of the enzymes, but without the first intermediate of the pathway, the enzymes will sit idle
doesn’t seem very efficient - since enzymes are being made but sit idle, bur advantage of this way of regulation is that when needed, the pathway is ready to go straight away.
Without the CAP site, can the gene still be expressed?
Yes.
The CAP site helps upregulate gene expression, but the RNA polymerase can still transcribe without the cap - just slower.
activator is not needed for gene expression to occur. It just increases efficiency.
What is tryptophan?
Activator, repressor, inducer, corepressor?
corepressor.
Feedback inhibition vs Feedback repression:
Feedback inhibition:
product of a pathway inhibits the activity of an enzyme earlier in the pathway
often end product binds to an enzyme for the first step, reducing its activity
Feedback repression:
where the product of a pathway interacts with a regulatory protein (e.g. repressor) to stop transcription of genes encoding enzymes in the pathway
end product is still going to be the signal the cell senses and uses to work out whether to express these genes or not.
e.g. (allosteric repressor protein inactive when Tryptophan unbound) Tryptophan (end product of pathway) binds to allosteric repressor protein to form active complex (corepressor-repressor). Binds to operator and blocks transcription.
Note: Here, Tryptophan is a corepressor, not an inducer (bc repressor is inactivated in presence of inducer but for feedback repression, repressor is activated by presence of corepressor)
Transcriptional attenuation:
Aside from regulation at promotor, regulation can also occur after transcriptional initiation
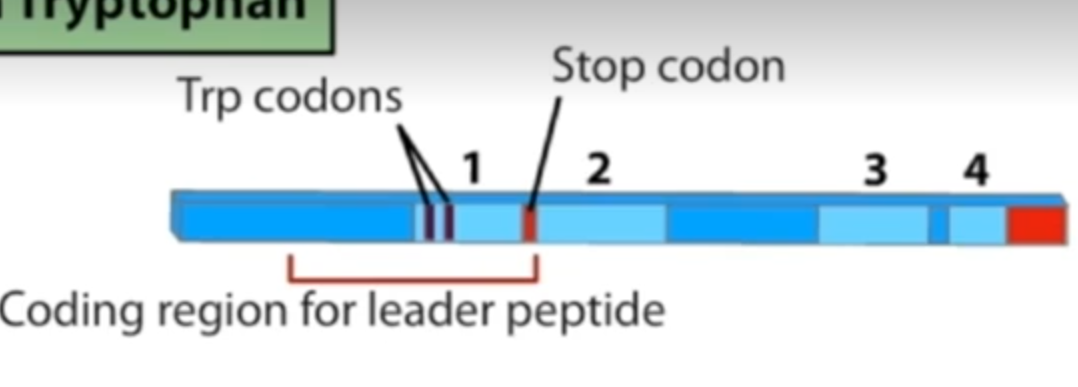
Involves an upstream leader sequence encoding a peptide (L, TrpL)
leader sequence located between operon and first structural gene
leader sequence has 2 consecutive tryptophan codons close to the start and an attenuation stem loop close to the end
Tryptophan absent = whole mRNA is transcribed
Tryptophan present = only leader sequence is transcribed
How does high/ low tryptophan affect transcriptional attenuation? (prokaryotes)
Refer to image:
The leader sequence:
The leader sequence has 4 distinct regions which can pair with each other (1 with 2 and 3 with 4, or 2 with 3).
when 3 and 4 pair, the following U-rich sequence is bound weakly, causing DNA and RNA polymerase to fall off.
there is a stop codon between regions 1 and 2
there are 2 trp codons towards start of leader sequence in region 1
High/ low tryptophan:
when the leader sequence is being transcribed, ribosome starts transcribing.
with high trp in cell, ribosome doesn’t pause until it reaches the stop codon. Stalled at this position, the ribosome covers regions 1 and 2, not allowing then to pair. So 3 and 4 pair, causing attenuation.
with low trp, ribosome stalls at trp codons bc there is not enough tRNA with Trp attached. Stalled here, regions 2 and 3 can pair. As 3 and 4 cannot pair, attenuation does not occur and RNA polymerase continues to transcribe the operon and a new ribosome translates the structural genes