Genetic Drift
1/38
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
39 Terms
H-W model
constancy of allele proportions from one generation to next.
Populations are not infinitely large, their sizes are not constant, so
fluctuations in allele frequency can occur by chance ( )
genetic drift
Systematic evolutionary forces can also change allele frequencies. What are those forces?
• Mutation
• Migration
• Selection
In real finite sized populations may result in changes to allele
proportions due to:
-segregation of alleles (Mendel’s First Law)
-variation in the no. of offspring between individuals
chance fluctuations in allele frequencies occur, particularly in
small populations, as a result of
random sampling of gametes
because small samples are frequently not representative, an
allelic frequency in the sample may
differ from that in the entire
pool of gametes
If # gametes in sample is 2N (where N = no. of individuals) the
probability that the sample contains exactly 0, 1, 2, 3....2N alleles of
type A is given by
successive terms of the expansion (pA + qa)2N where
p and q are the allele frequencies of A and a in the parental generation
and p + q = 1
Probability sample contains exactly i alleles of type A is
[(2N)!/i!(2N-i)!] piq2N-i (binomial probability)
genetic drift causes
random changes in allele frequencies. alleles are lost from the population
the direction of random changes is neutral:
no systematic tendency
for the frequency of alleles to move up or down
Genetic drift is a===== process
random
the outcome of genetic drift cannot
be stated with certainty
genetic drift removes
genetic variation
The
probability that an individual chosen at random from the
population is heterozygous after t generations of random mating is
where H0 is the initial probability of being a heterozygote
and N is the population size
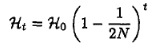
The probability of fixation of an allele, say A, (i.e. pA = 1) is equal to
the frequency of the A allele in the starting population
Genetic Drift
Genetic drift affects evolution in two ways:
• the dispersive force removes genetic variation from
populations (removal is inversely proportional to the
population size, so genetic drift is a very weak dispersive
force in most natural populations).
• drift’s effect on the probability of survival of new mutations,
an effect that is important even in the largest of populations.
• probability of fixation of an allele (i.e. pA = 1) is equal to
the frequency of the A allele in the starting population
The dispersive aspect of genetic drift is countered by
mutation, which
puts variation back into populations. These two forces reach an
equilibrium and can account for much of observed molecular variation.
Mutation provides
he raw material for evolutionary change but by
itself is a very weak force for changing allele frequency (in SNPs)
The mutation must be
non deleterious otherwise selection will
remove it from the population
Neutral Theory (1968 Kimura):
most polymorphisms observed at the
molecular level are selectively neutral. They produce such small
effects on ability of organism to survive and reproduce that they are
completely equivalent in terms of natural selection. (Their fate is
determined by genetic drift)
Mutation followed by genetic drift produces the variants
observed in
STRs
Mutation rates are high in STRs
(~10-3 per locus per
generation compared to SNPs (2.5 x 10-8 per base per
generation)
High mutation rate in STRs are why they are
highly polymorphic
STR mutational mechanism:
called DNA slippage, polymerase slippage, or slipped
strand mispairing (Kornberg 1964)
DNA slippage Looping out of
extending strand
+1
DNA slippage Looping out of
template strand
-1
Stepwise mutation model (SMM)
alleles can mutate up or down by one (or occasionally a small no. of
repeat units)
Factors Influencing STR Mutation
• No of Repeats: rate increases with repeat number
• Repeat unit size: dinucleotide repeats have higher mutation rate than
trinucleotide repeats
• Base composition of repeats: repeat units with a high AT content
mutate faster than those with a high GC content (template stability
affect?)
STR Mutations
Factors Influencing STR Mutation (con’d)
• Flanking sequence: if near hotspots of recombination,
mutation increases
• Sex: mutation of male gametes > female (more DNA
replication in males).
• Age: older males, more mitoses, more mutations than
younger males
• Interruptions in the STR repeats: more interruptions, lower
mutation rate
13
Migration, also known as gene flow, is
any movement of
individuals, and/or the genetic material they carry, from one
population to another (ie the movement of alleles from one
area (deme, population, region)
includes various events, such as pollen being blown to a
new destination or people moving to new cities or
countries
In a subdivided population random genetic drift results in
genetic divergence among subpopulations.
Migration
(movement of individuals among subpopulations) limits the
amount of genetic divergence that can occur.
If migrating individuals stay and mate with the
destination individuals,
they can provide a sudden influx
of alleles.
After mating is established between the migrating and
destination individuals, the migrating individuals will
contribute gametes carrying alleles that can alter the
existing proportion of alleles in the destination
population.
The genetic variation in modern human populations has
been critically shaped by
gene flow.
gene
flow introduced new genetic variation to the human
population through
interbreeding between ancient
humans and Neanderthals
Rates of migration typically much greater than
rates of
mutation, changes in allele frequency occur much
faster with migration
Natural selection
differential survival and
reproduction of
individuals due to
differences in phenotype