BIOC Slides 2 (Amino Acids + Proteins)
1/84
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
85 Terms
What are proteins and their characteristics?
linear polymers built out of a.a
a protein’s final 3D structure depends on its sequence of a.a
proteins can interact with each other and other molecules to form complexes
ex. hemoglobin → combo of 4 different polypeptides
Proteins can be rigid or flexible
How many amino acids are there in living things and how are they used to make different proteins?
there are 20 key amino acids in living things
you can vary the a.a by caring the side chains (R)
aliphatic
a compound with an open chain structure (alkane)
What are the non-polar and aliphatic R groups for amino acids?
Glycine (Gly, G)
Alanine (Ala, A)
Valine (Val, V)
Methionine (Met, M)
Proline (Pro, P)
*all of these are also hydrophobic
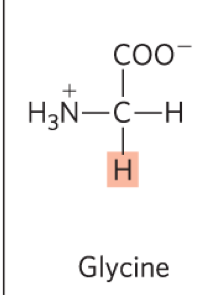
Describe Glycine:
What is the abb.?
structure?
features?
Gly, G
Features:
the simplest a.a, R group is H
the only archiral a.a
technically not really alipathic or hydrophobic (but closet catergory, so we put it here: non-polar and aliphatic)
Describe Alanine:
What is the abb.?
structure?
features?
Ala, a
Features:
contains a methyl group
Describe Valine, Leucine, and isouecine:
What is the abb.?
structure?
features?
Valine: Val, V
Leucine: Leu, L
Isouecine: ile, I → i
Features:
all contain hydrocarbon side chains (R)
ile also has a second chiral carbon
Describe Methionine:
What is the abb.?
structure?
features?
Met, M
Features:
also has a hydrocarbon side chain (R), expect it has a non-polar thio-ether (C-sC) group
hydrophobic R group
Describe Proline:
What is the abb.?
structure?
features?
Pro, P
Features:
has an aliphatic side chain with a twist
end of the R groups is bound to the N of the amino group
this forms a 5 membered ring
it is not aromatic
ring structure makes the a.a. more restrained
introduces kinks into a.a chain (i.e polypeptides)
What do all the non-polar and aliphatic R groups for amino acids have in common?
all of these amino acids are hydrophobic and tend to cluster together
different sized and shaped R groups allow for close packing
they are usually found in the center of a protein away from water
driven by the hydrophobic effect (ie. increased entropy when hydrophobic molecules cluster together)
usually not reactive
Describe features of all aromatic R groups:
contain aromatic ring (phenyl rings)
participate in hydrophobic interactions
What are the amino acids with aromatic R group
Phenylalanine
Tyrosine
Tryptophan
Describe Phenylalanine:
What is the abb.?
structure?
features?
Phe, P
Features:
contains a hydrophobic phenyl ring
Describe Tryrosine:
What is the abb.?
structure?
features?
Try, Y
Features:
similar to Phe, execpt it has a reactive polar OH group which can H-bond
Describe Tryptophan :
What is the abb.?
structure?
features?
Try, W
Features:
contains an indole group
indole group: a 5 membered ring connected to a 6 membered group
this is aromatic
NH is reactive and can H bond
What are the positively charged R groups in amino acids?
Lysine
Argine
Histidine
These are basic!
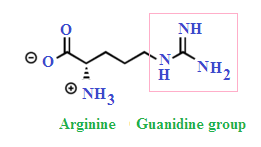
Describe lysine and arginine
What is the abb.?
structure?
features?
lysine: Lys, K
arginine: Arg, R
Features:
contain long chains with ionizable groups
Lys = amino group
Arg= guanidinium group
they are both postively charged at pH (pka >/= 10)
Describe Hisitidine:
What is the abb.?
structure?
features?
His, H
Features:
also has an ionizable group (imidazole ring) with a pka 6
thus, pure His at pH 7 will have an unchanged imidazole ring
imidazole ring: a 5 membered ring with N on 1 and 3
however His in protein will often have altered pka, closer to pH 7 and exist as a mixture of its acid (pos. charged) and c. base (unchanged forms)
this makes is a good proton donor and proton acceptor
this means H can act as a buffer (acid-base catalyst)
it can be charged or uncharged depending on its location
often found in the active site of enzymes
What do all negatively charged R groups have in common? - amino acids
contain carboxyl groups as R-groups
negatively charged at pH 7 and pka is below 4
acidic
What are the negatively charged R group amino acids?
Aspartate
Glutamate
Describe Aspartate and Glutamate:
What is the abb.?
structure?
features?
Aspartate: Asp, D
Glutamate: Glu, E
Features:
Glu is 1 carbon longer than Asp
remember “-ate” at pH 7 but “-ic acid” is strong acid
charged amino acids are often found on the surface of proteins where they interact with water away from the hydrophobic a.a
Describe what Polar R groups all have in common?
uncharged R groups at pH 7
can H bond, more hydrophilic
What are the amino acids with polar R groups?
cystine
asparagine
glutamine
Describe cystine:
What is the abb.?
structure?
features?
Cys, C
Features:
contains a sulfhdryl or thiol (SH) group
is polar and weakly H bond is reactive
can form disulfide (covalent) bonds
this can link 2 parts of a chain or 2 separate chains together
done by the oxidation (loss of e-) of 2 cysteine residues to cystine residues (non-polar)
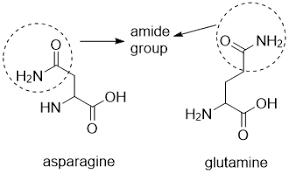
Describe Asparagine and Glutamine:
What is the abb.?
structure?
features?
Asparagine: Asn, N
Glutamine: Gln, Q
Features:
are detrivatives of asparate and glutamate
contains a terminal carbonyl amide
terminal amine is usually uncharged
What is a primary structure?
linear sequence of amino acid linked by peptide bonds to form a polypeptide
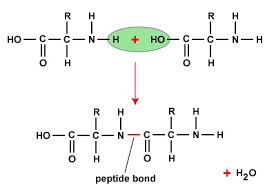
what is a peptide bond?
linkage of an alpha-carbonyl of one amino acid to the alpha amino group of another and the loss of H20.
the formation of a peptide bond isn’t energetically favourable but once it is formed it is stable and a high activation energy would be need to break the peptide bond
What is a polypeptide?
is a series of amino acid residues linked by a peptide bond
What is a residue?
an amino acid unit in a polypeptide
What does it mean when you say polypeptides have polarity?
one end has a free amino (NH3+) (left side)group and one has a free carboxyl group (COO-) (right side)
A polypeptide consists of a backbone of repeats with variable side chains
the backbone is polar (hydrophilic) and rich in H-bonding potential
therefore all the carboxyls and amines can H bond with the exception of proline, which has limited H bonding ability.
how many amino acids residues are typically in a protein?
the polypeptide chain can contain 50-20,000 amino acid residues
How is a protein’s molecular weight measured?
Daltons or kilodalonts (kDa)
1Da = 1g/mole
What does the primary sequence of amino acids allow us to know?
Determine shape
Determine the function ex. catalyst function of enzyme
understand diease ex. cystic fibrosis
understand evoluntary history
Point mutation nomenclature

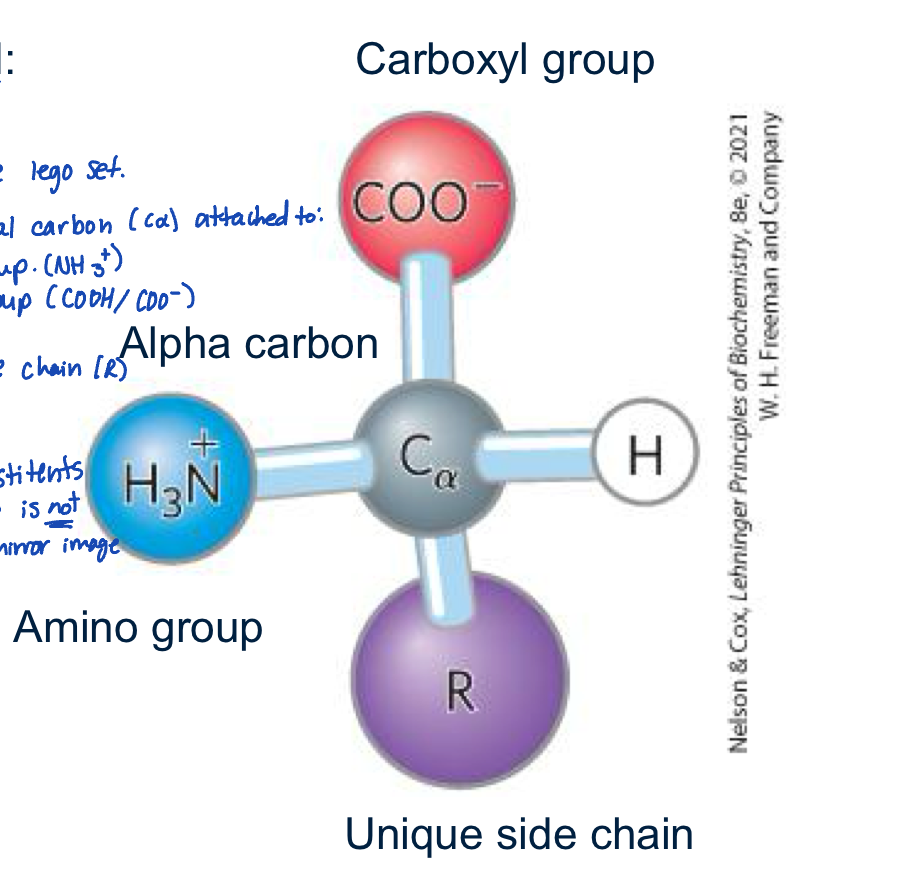
What is the structure of an alpha amino acid?
is like the ultimate lego set
contains an essential carbon (c-alpha) attached to:
an amino group
carboxyl group
hydrogen
unique side chain R
note the c-alpha is chiral
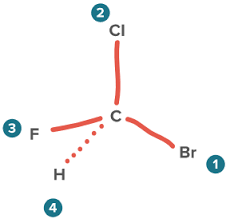
what is a chiral center?
an atom with its substituents arranged so the molecule is not superimposable on its mirror image
for each amino acid there are 2 enantimors expect glycine
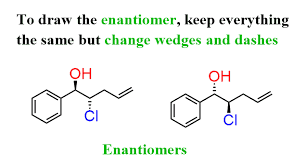
enantimor
a pair of molecules with atleast 1 chiral center that are mirror images of each other
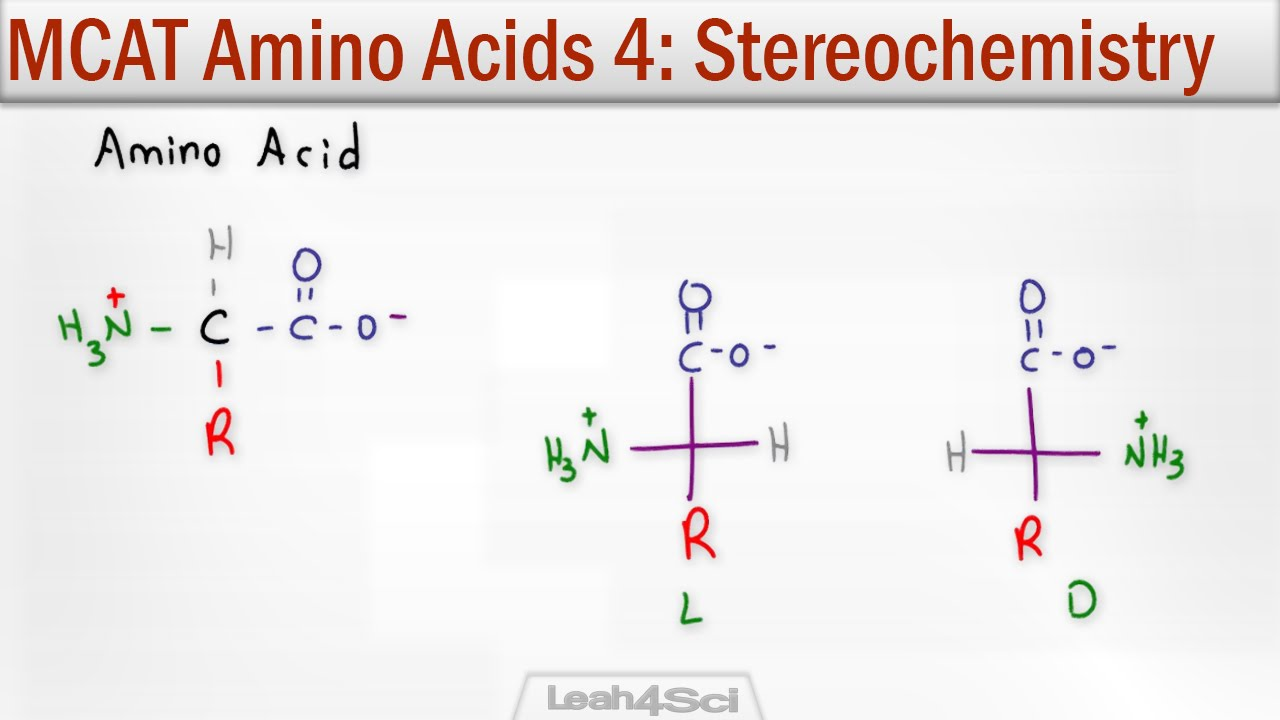
L and D configurations and which one is more common in nautre?
L is more common in nature
Zwitterionic form
ions with both pos and negative charge but over all net charge is zero.
Why are polypeptides conformally constrained?
The peptide bond has a double bond characteristic
this is because of resonance between the peptide and the carbonyl
as a result the peptide bond is planar
this is turn locks a series of atoms into a plane
so there are not rotations about the peptide bonds
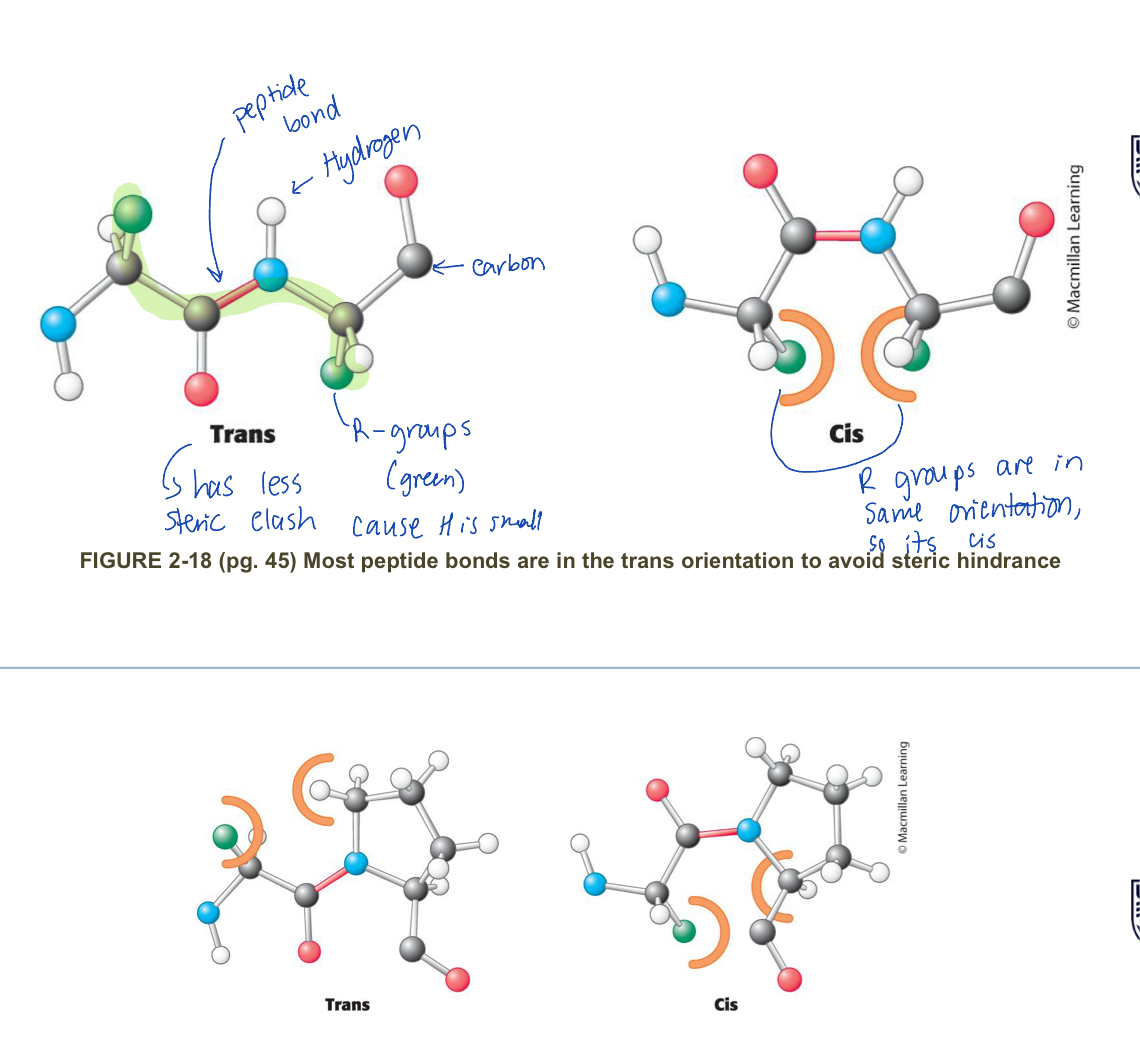
what orientations do double bonds exist in and how does that impact peptide bonds?
Double bonds exist as:
cis
trans
therefore:
peptide bonds can only be cis or trans, since they act like double bonds
however, because of steric hinderness, all peptide bonds are trans expect for x-pro peptide bond where both cis and trans occur
Why are peptide bonds flexible?
the bond between N-C(alpha) and the bond between C alpha CO are free to rotate
this provides flexability, allowing the backbone to fold in many ways
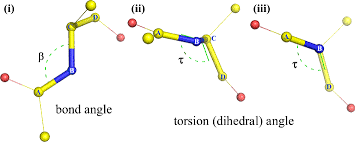
dihedral angle
how we measure the amount of rotation about the bond
this ranges from -180 - + 180 degrees
note: this is different from bond angle
bond angle: a bend in the bond between molecules
dihedral angle: how the molecules rotate/twist due to the bond
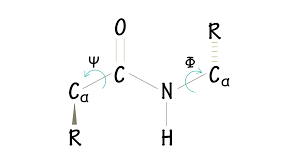
Phi and psi bonds?
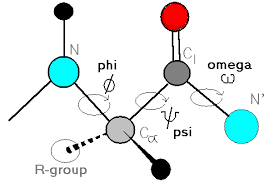
how to find the N-C alpha angle?
the N-C alpha angle = Phi + psi
Can all combinations of phi and psi be formed? why or why not?
no, not all combos are allowed due to steric hinderness
this further limits the number of structures a protein adopts
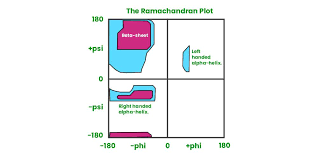
The possible combos are shown in a Ramachandran plot
Describe a Ramachandran plot
areas of dark colour = very favourable
areas of light colour = less favourable, but possible
white areas = not permitted
why are random coils formed?
large molecules that can freely rotate among many bonds will assume random coils (i.e. a mix of different structures)
why do proteins fold into a single structure?
because proteins have a series of limitations on what orientations they can adopt (i.e the planar peptide bond and limitations on dihedral angles) they can often spontaneously fold into a single structure.
Secondary structure of proteins
is the spatial arrangement of amino acid residues in a polypeptide that are relatively close to each other in a linear sequence (alpha helices and beta sheets/strands)
alpha helix
polypeptide backbone forms the inner part of a right-handed helix with the side chains sticking outwards
the helix is stabilized by intra-chain H bonds between the NH and CO groups of the backbone
Characteristics of alpha helix?
the R groups in an alpha helix are almost perpendicular to the axis of a helix
it has dihedral angle of phi =-60 degrees and psi = - 45 degrees
the c= o of residue i forms H-bonds with the N-H of residue i+4 (4 residues further)
all the N-H and C=O in the backbone are H-bonded expect at the ends
each amino acid residue in the helix increases the helix length by 1.5 A (helix rises by 1.5A per a.a.)
the R-groups of i, i+1, and i+2 point in different idreactions
the R-groups of i, i+3 and i+4 point in similar direactions
the helix is almost always right handed in proteins
Left handed helices are permitted but rare (not as stable)
ALpha helices are shown as twisted ribbons or rods
What is the alpha helix content in proteins
vaires
some have no alpha helixes and other proteins are all alpha helics
2 alpha-helices can interwine into coiled colds
usually alpha-helics are less than 45 A

is peptide formation favourable? why does peptide formation occur?
peptide breakage is more favourable than formation
but acitivtion energy makes it hard to break
What is a beta-sheet/strands?
usually polypeptide strans from the same molecule
associated as stacks or chains in an extended zigzag
stabilized by interstrand H-bond between N-H and C=O groups
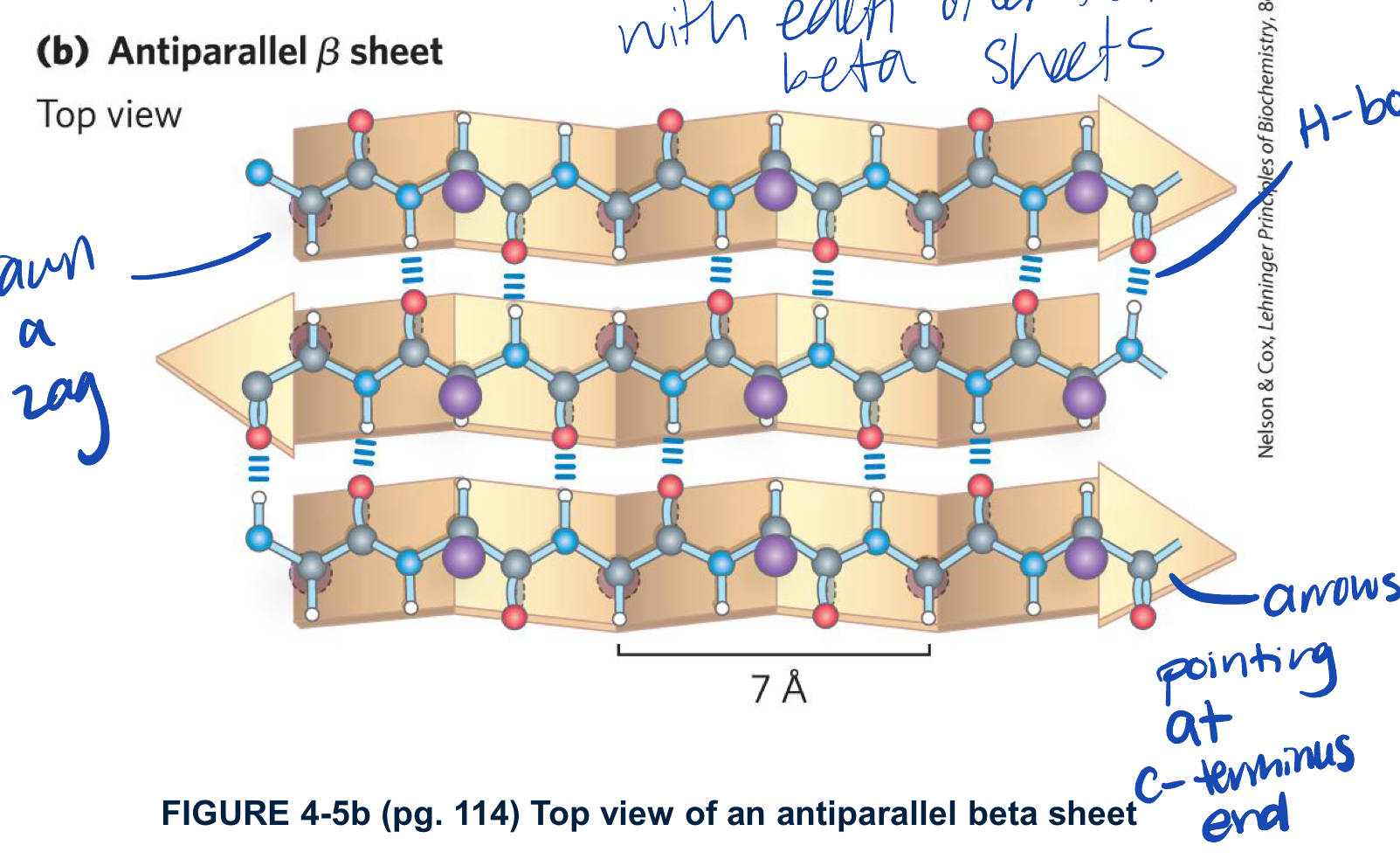
Describe anti-parallel B-sheet/strand structure
fro each amino acid extends to B-streand by 3.5A (more spread out than alpha helix)
the R-group of adjacent residues point in oppsite direactions perpendicular to the plane of the strand or sheet
the strands are organized into sheets
the N-Hand C=O of a single residue i on one B-strand H-bonds to a singla residue i on the other B-strand oppsite
has dihedral angles of phi -139 degrees and psi +135 degrees
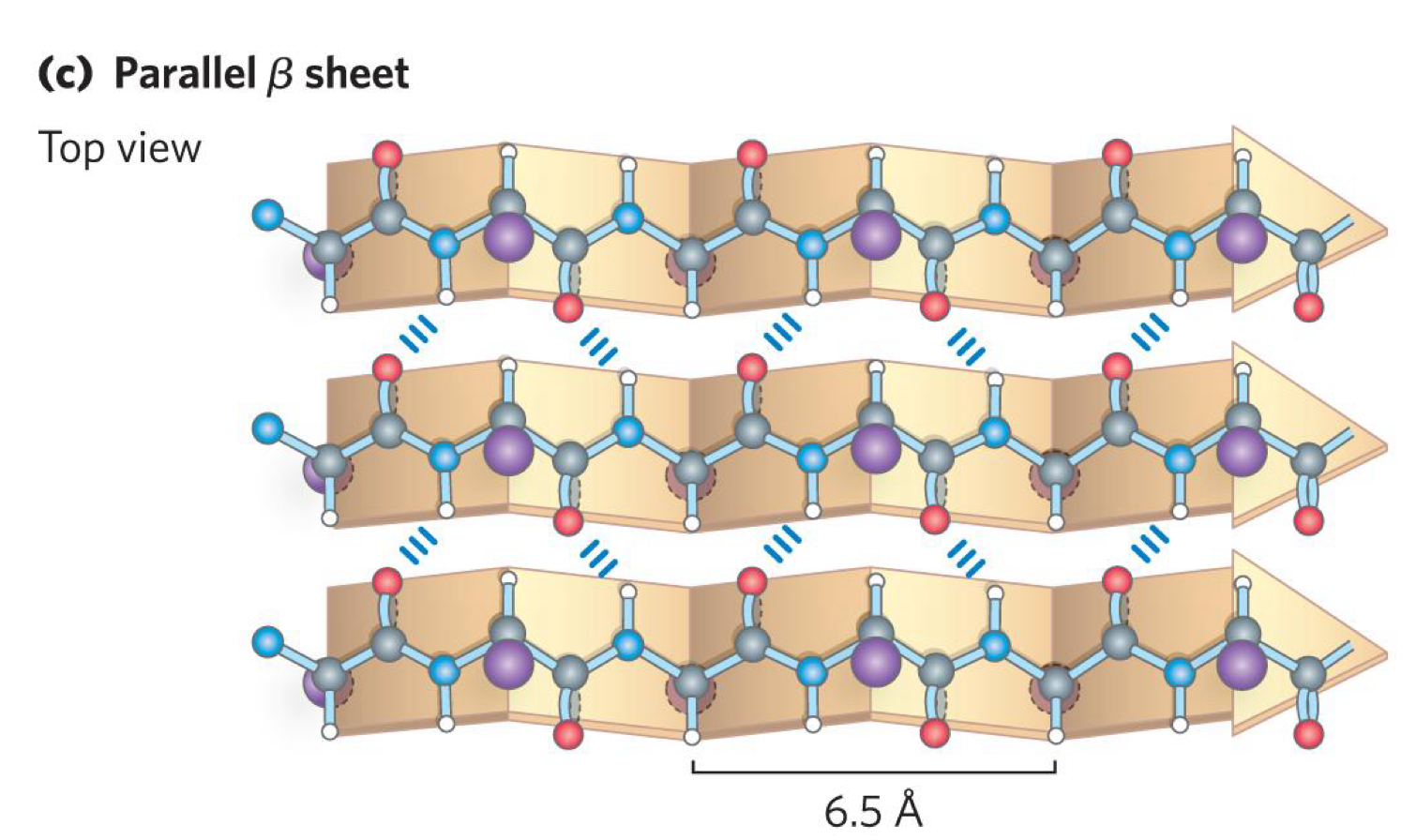
describe parallel B-sheet/strand structure
like anti-parallel B-sheet expect B-strands run in same direaction
each residue in the B-strand only extends the strand by 3.25 A
have different dihedral angle phi = 119 degrees and psi +113 degrees
the N-H of residue i in one B-strand H-bonds to residue i in the other B-strand
C=O of residue i in the first strand H-bonds to N-H group of i+2 in the second strand
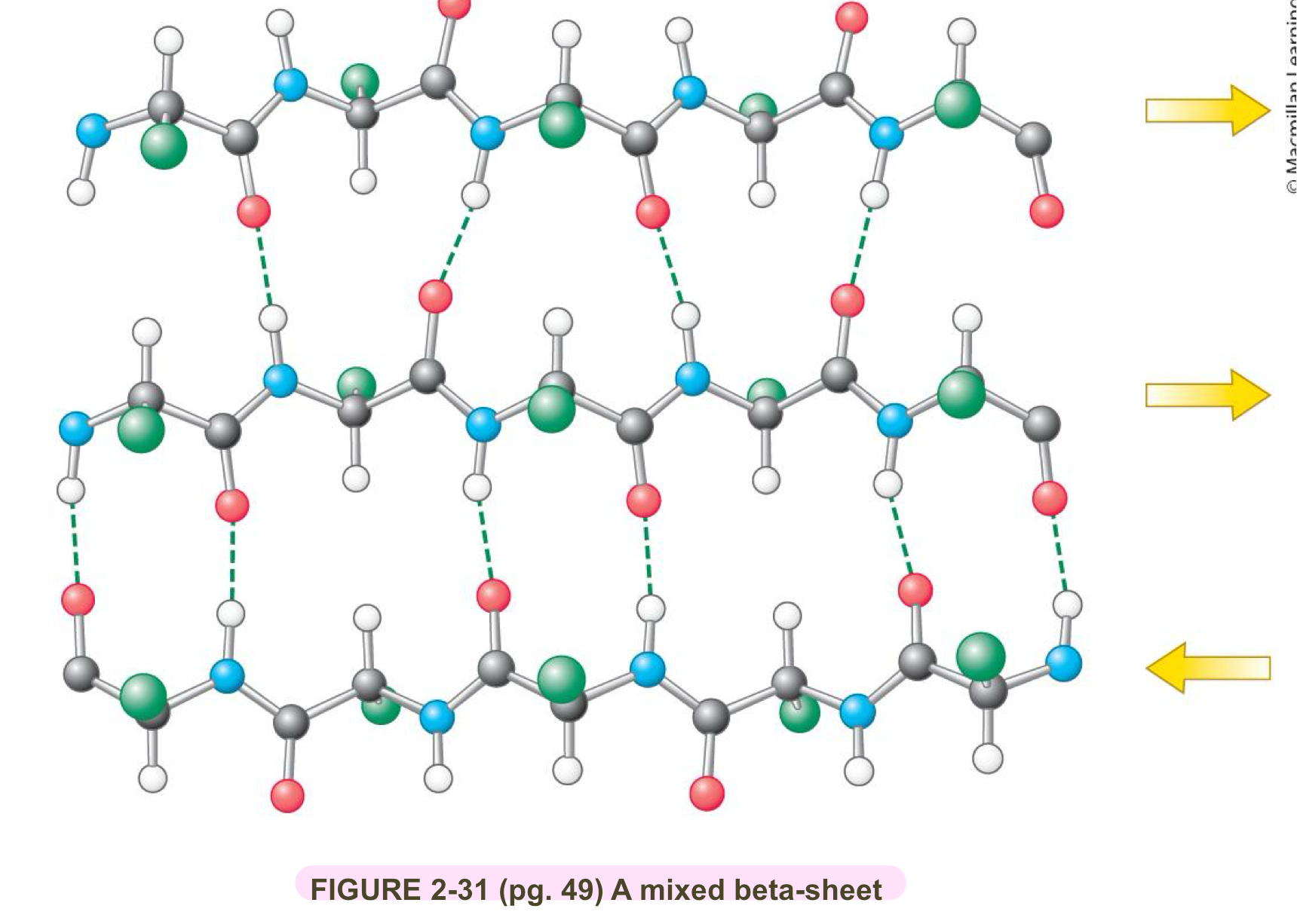
Describe general B-sheet and B-strands characteristics
B-sheets can be mixed (i.e both parallel and anti-parallel B-stands)
B-sheets cna be flat or twistied
the distance between B-strands in primary strucutre a.a can be small or large
B-strands can be large or small
B-strand can be shown as borad arrows pointing to C-terminal end
both can be built on itself to form a beta barrel
what are loops and turns
non-repetitive secondary structure that connects and changes directions of regular repeating elements like alpha helix and B-sheets
loops are generally longer and more flexible, and have less H-bonding
wide range of phi and psi
turns are shorter, tight, stretches stabilized by internal H-bonding
Prolines are common to give tight angles
Glycine accommodates angles incurred by proline
Tertiary structure
the spatial arrangement of a.a. residues that are far apart from each other in linear sequence as well as the pattern of disulphide bonds
the polypeptide’s 3D structure
each protein’s tertiary structure is different
is limited to a single polypeptide that is folded into a structure
in most 3 structures, the dihedral angles for each residue fall into permissible areas (blue areas) of ramachran plot.
Myoglobin
example of Tertiary structure
oxygen storage protein in mammalian muscle
single polypeptide chain of 153 aa residues
contains a heme group (iron in protoporhyin ring) where the oxygen bind
70% of the chain is in alpha helices (total of 8 helices in protein)
the rest are largley loops and turned
the core of the protein is almost exclusively composed of hydrophobic residuces expect for 2 His residues which is needed by heme
the surface is composed of molar polar/charged residues (some non-polar)
myoglobin binds oxygen with high affinity and only releases it when the [O2] is really low
structural domains
some proteins have multiple compact structures called domains linked by flexiable sections in the polypeptide these often have no defined sturcture
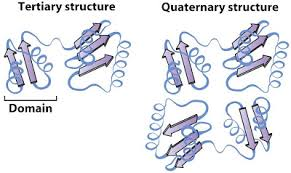
Quaternary structure
the spatial arrangement of multiple folded structures (folded polpeptides) and the nature of their interactions along with disulfide bonds between subunits
some proteins are composed with more than 1 folded polypeptide (subunit)
Homomer vs heteromers
Homomer: subunits that are identical
heteromers: subunits that are different
multimer
2+ monoers combined
some proteins must be monoers to survive
Protomer
the base unit in a quat. structure (the basic structural subunit that repeats to form a larger protein complex)
usually repeptive (but not always) monomeric (composed of a single unit) in nature
But the structure is not the same as a monomer
Hemoglobin
example of quat. structure
oxygen transporter in mammals
composed of 4 subunits (dimer of dimers)
2x alpha subunits (alpha-globin)
2x B-subunits (B-globin)
Hemoglobin (Hb) cannot function unless it is a tetramer
the protomer for Hb is an alpha-B-dimer
What drives protein folding?
folding is driven by thermodynamics
finding the most stable complex, which is the most neg. delta G
the difference in free energy between folded and unfolded is a small difference
~20-60KJ/mol
mostly driven by entropy
the hydrophobic redsidues are excluded from water in the core whilst the hydrophobic residues are on the surface
How do we predict protein’s 3D structure
AI programs like AlphaFOLD2 or RossetaFold predict protein structure based one primary structure
Why must the polypeptide have H-bonding?
in order to bury the polar/hydrophobic bakbone of the polypeptide in the core it needs to H-bone
How can alpha helix structures get destablized?
if unpaired, charged, or polar groups are in the hydrophobic core the alpha helix will destabilize
Are alpha helixes and B-strands hydrophobic, hydrophilic, or amphipathic?
amphipathic
Can a portion of the primary seq. define secondary sequence?
Yes and no
certain a.a. residues are more likely or less likely to be found in and stabilized (or destabilized) alpha helices and B-strands/sheets
experiments have shown that the exact same portion of a sequence in 2 different proteins can adopt different secondary structures
therefore, we can’t always determine secondary structure by looking at a portion of the primary sequence
the overall tertiary structure is influenced by the secondary structure
Which amino acids are considered alpha helix wreckers?
pro
gly
and to a lesser extent B-strand wreckers
What are 3 characteristics of protein folding?
folding tends to be an all or nothing process (usually either folded or not)
It is co-operative as one portion of the protein folds (eg. B-sheets)
it will influence how another portion forms
don’t need to sample every structure
there are usually many possible pathways so we depict this as a free energy funnel
In an unfolded protein state, there are many possible structures with high free energy, but as these species fold, the free energy decreases until you reach the folded state.
are all proteins a 3D structure?
not all proteins have a 3D structure
some proteins only fold into a single structure when they bind something
some proteins are in equilibirum between 2 sturcutres
What are 3 methods to determine the 3D structure of proteins?
x-ray crystallography: uses x-rays to measure electron density (this was how structure of myoglobin and hemoglboin were determined)
Nuclear magnetic Resonance (NMR): measured the location of nuclei
Cryo EM: uses a beam of e- to visualize a frozen protein
What are the different types of post transitional modification?
Phosphorylation
Glycosylation
Hydroxylation
carboxylation
Acetylation
Methylation
Phosphorylation
the attachment of phosphate group usually OH of an R-group (ser,Thr, Tyr)
to activitate or inactive a protein
Glycosylation
the attachment of 1 or more sugars to a residue (asn, Thr, Ser)
common is surface lavelling most proteins on cell surface are glycosylated
Hydroxylation
addition of OH group
usually to Pro
ex. fibre stabiliztion in collagen in skin, if someone has scurvy it could breakdown
Carboxylation
add a carboxyl group
usually to Glu
important in clothing
Acetylation
addition of an acetyl group to an amino group
Lys, Arg
Methylation
addition of a methyl group to an amino group
Lys, Arg
most proteins are cleaved or trimmed after synthesis
this could activate or deactivtate them
ex. fibrogen gets cut into fibrin (active form)
important for blood clotting but don’t want clots all the time
multiple proteins can be formed from a single long polypeptide
ex. virus
Protein folding as a funnel
as you go down the funnel it is more stable
sometimes proteins can get trapped because it takes alot of energy to unfold and refold into something more stable (makes chaperone proteins important)
