micro to macro - amino acids to proteins
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
What are amino acids and the basic structural features of amino acids?
building blocks of proteins
3D molecules
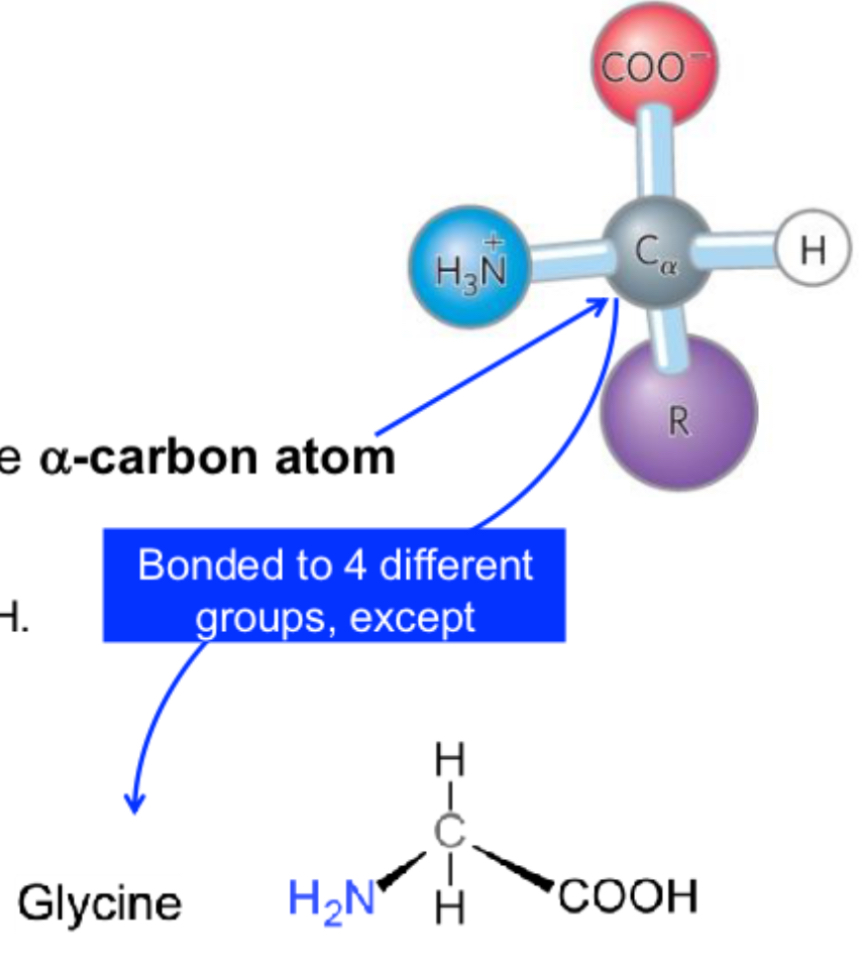
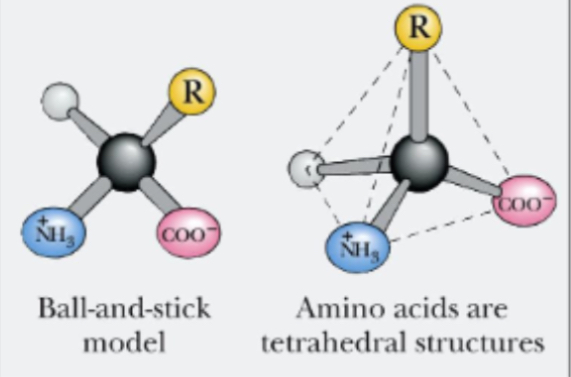
4 groups arranged tetrahedral around alpha - carbon atom: -H, NH2, COOH, R
Glycine is simplest amino acids with R- as hydrogen
What defines an alpha amino acid and which amino acids are proteinogenic?
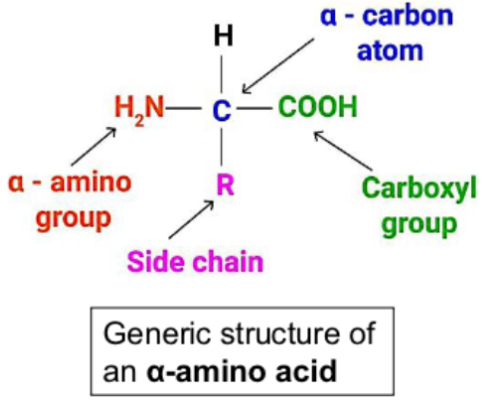
alpha amino acids have amino and carboxyl group attached to same central sp3 - hybridised carbon
All amino acids that constitute of proteins and are coded in genome are alpha amino acids aka proteinogenic amino acids
All proteins essentially composed of 20 naturally occurring alpha amino acids. Some proteins however dont contain all 20 amino acids.
Draw a general alpha amino acids
Can you name the 20 amino acids and their side chains?
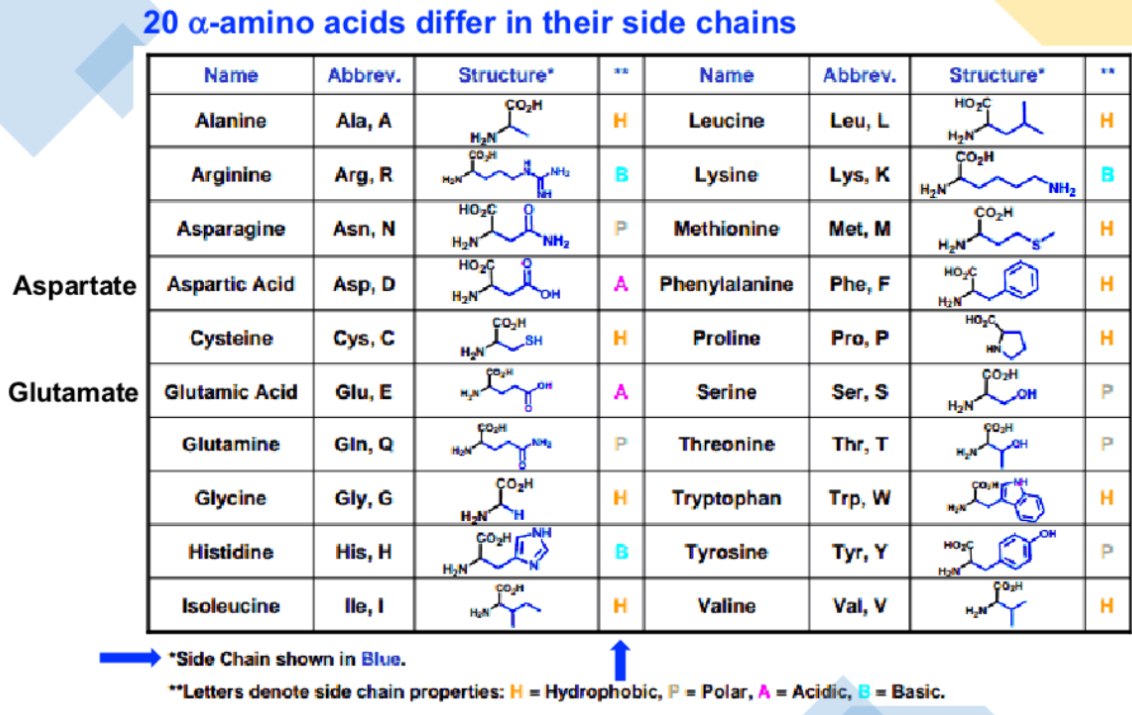
Whats a beta and gamma amino acids?
Beta= NH2 attaches to beta carbon which is 2nd carbon atom away from carboxyl group.
So NH2-CH2-CH(R)-COOH
Gamma = NH2 attached to gamma carbon which is 3rd carbon away from carboxyl group
So NHS-CH2-CH2-CH(R)-COOH
What are non proteinogenic amino acids?
not directly incorporated into proteins during process of translation in living organisms.
Occur naturally but not used in proteins or modified forms of proteinogenic amino acids
Whats a primary and secondary amine?
primary - 1H of ammonia replaced by 1 alkyl group. R-NH2. So N bonded to 1 carbon and 2H
Secondary - 2H of ammonia replaced w/ 2 alkyl groups. R-NH-R2. So N bonded to 2C and 1H
Why is glycine special amongst amino acids?
Glycines side chain is just a hydrogen atom so it’s not chiral. It doesn’t have a L or D form
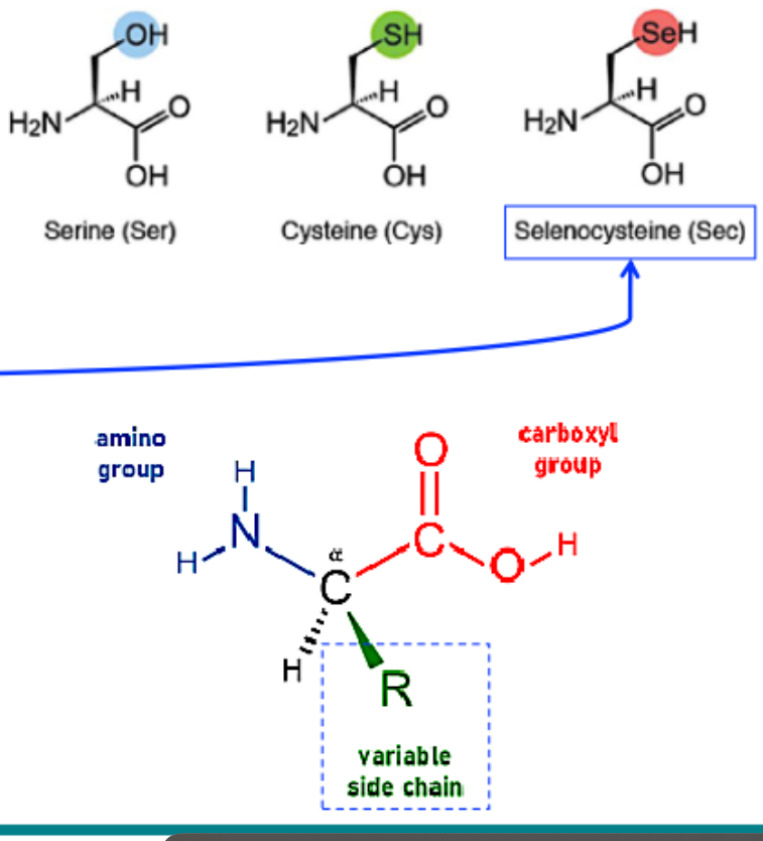
What is selenocysteine and why is it considered the “21st” amino acid?
its an AA that contains selenium and some enzymes like glutathione peroxidase, formate dehydrogenase contain selenocysteine.
Its recognised in some species and functions similarly to cystine but w/ selenium instead of sulfur
How do side chains - R groups - determine amino acids?
Side chains determine:
chemical properties (polarity, charge, hydrophobicity, hydrophilicity)
Their reactions (spontaneous or enzyme catalysed)
How AA interact (inter/intramolecular forces)
Folding, reactivity and overall protein structure/function
How are amino acids be classified?
Based on side chain R:
Non polar. E.g alkyl chains, hyrophobic
Polar (uncharged) e.g amides, alcohols
Acidic and polar e.g carboxylic groups and phenols
Basic and polar e.g amines
Can also be classified on nutrional/physiological roles:
essential AA - can’t be synthesised by humans, must obtain from diet
Non essential AA - can be synthesised in body
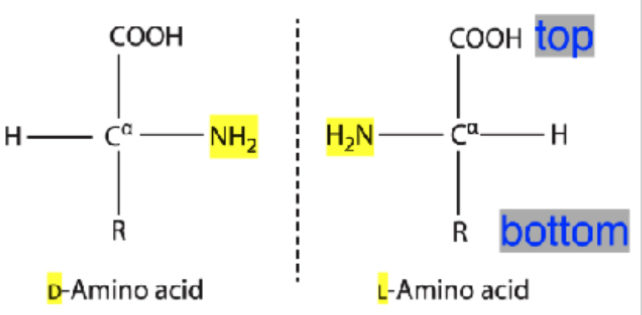
Which stereoisomer form (L or D) of amino acids are found in living proteins and why?
virtually all AA are L-form in living organisms.
Human proteins contain only L-amino amino acids (and glycine)
Some exception include - cone snail toxins - incredibly toxic venom containing D- amino acids and some fungal Ab contain D-amino acids
What does an L and D amino acid look like?
L- NH2 ON left
D - NHS ON right
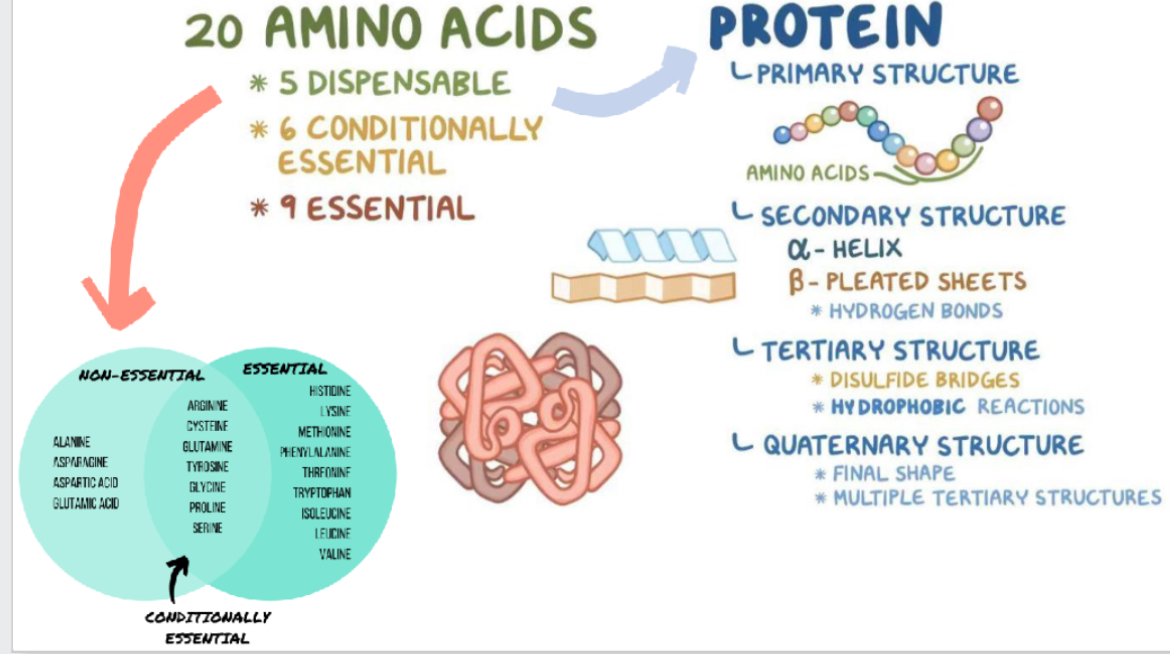
How many essential and non essential amino acids are there and what 4 structures is protein made of?
9 essential
11 non essential
Primary, secondary, tertiary , quaternary
What 3 sources do carbon skeleton precursors derive from?
Glycolysis (light red)
Citric acid cycle (blur)
Pentose phosphate pathway (purple)
These feed into pathways for synthesising non‑essential amino acids or precursors for essential ones.
Humans can only synthesize 11 non essential AA
Some AA are precursors of other AA
Also AA are synthesised from common metabolic intermediates
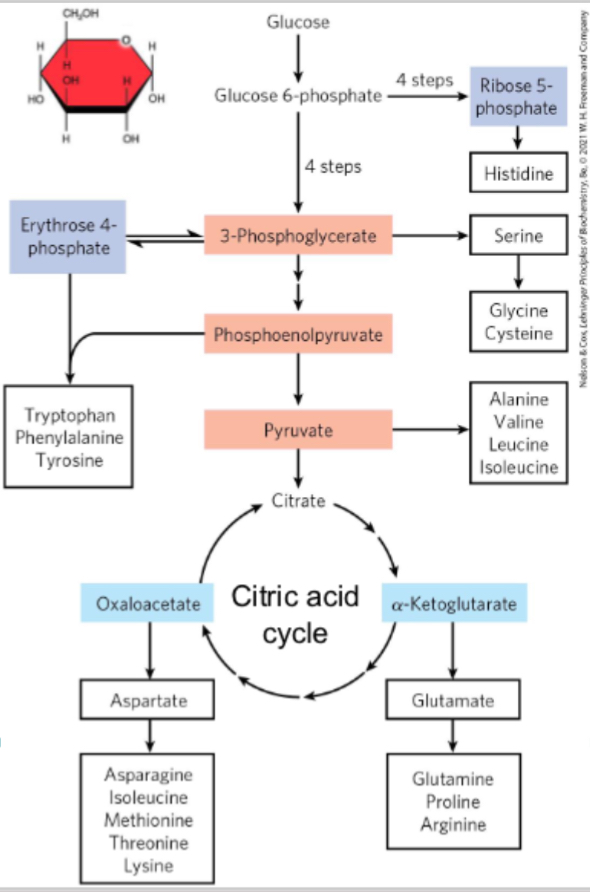
How do humans typically make non-essential amino acids?
Humans use transamination: transferring an amino group from one amino acid to an α-keto acid (a metabolic intermediate) to form a new amino acid. That works for amino acids whose keto-acid precursors are common intermediates.
Why can’t humans synthesize essential amino acids via transamination like microbes or plants can?
Because in humans, the α-keto acids that would lead to essential amino acids are not part of common metabolic pathways. Microbes and plants have additional enzymes to build those precursors; humans lack those enzymes. Plant and microbes alpha keto aciids are common intermediates (enzyme needed for them are lacking) so transamination isn’t an option. But theyre present in common pathways of microorganism and plants
What are the amino acid precursors?
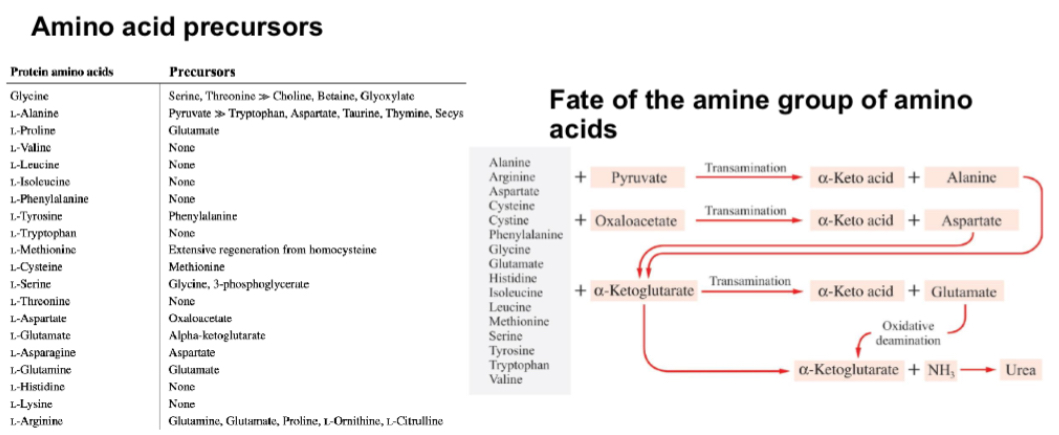
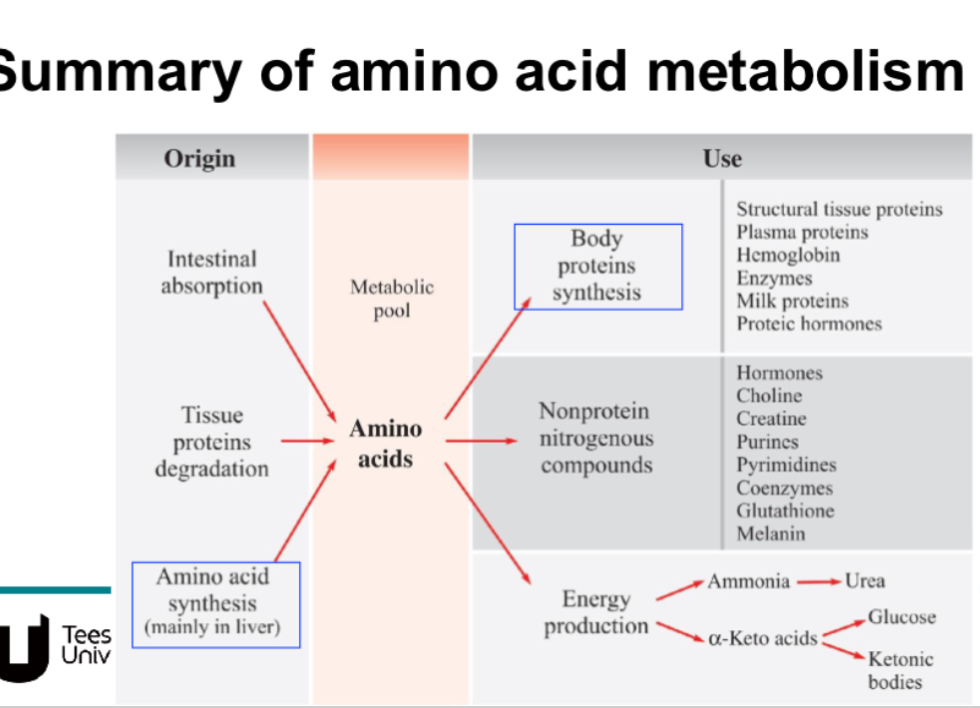
Summary of amino acid metabolism? Whats its original and its use?
What are the main steps in gene → protein?
Transcription: in nucleus, DNA → pre‑mRNA → mature mRNA, which exits nucleus
Translation: in cytoplasm, ribosome reads mRNA codons, tRNAs bring amino acids, polypeptide chain forms
The three phases of translation: initiation, elongation, termination.
It needs ribosome, tRNA, free AA
After translation, how is a protein folded and processed?
The newly formed polypeptide must fold into correct 3D structure
Post-translational modifications (e.g. acetylation, phosphorylation, cleavage) may occur
Chaperone proteins often help folding
Misfolding can lead to disease (e.g. amyloidosis, prion diseases)
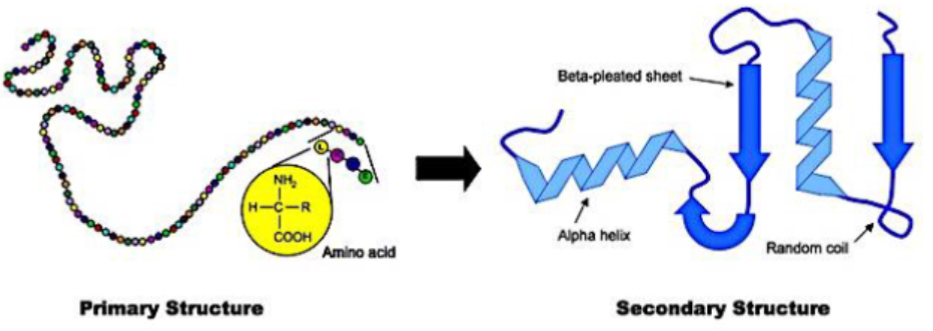
What defines primary structure of a protein?
The linear sequence of amino acids, forming backbone of protein. joined by peptide (amide) bonds via condensation reactions to form polypeptides, ribosome condensed 2 amino acids into dipeptide forming peptide bonds. Bonds formed covalent
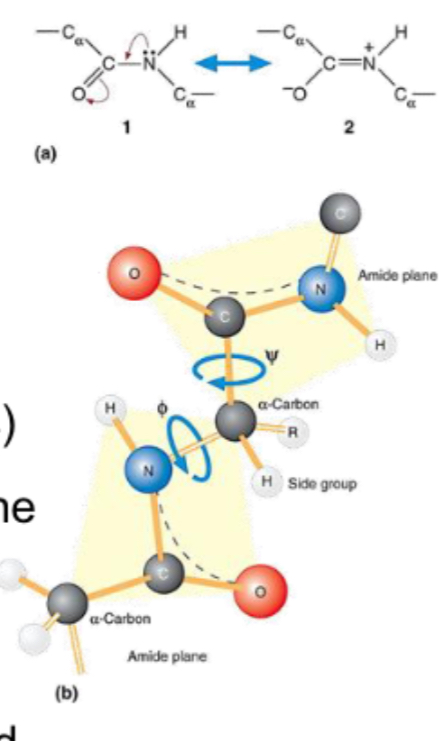
What is a peptide bond and what special properties does it have?
A covalent bond between the carboxyl carbon of one amino acid and the amino nitrogen of another. It has partial double-bond character (resonance), making it rigid and planar. As a result, rotation around this bond is limited so limits number of conformational possibilities; the side chains (R groups) can rotate.
What are the major types of secondary structures in proteins? And what is secondary structure?
Alpha helix: a spiral structure stabilized by hydrogen bonds between backbone CO and NH groups
Beta sheet: strands of peptide backbone lying side by side (parallel or antiparallel) held by hydrogen bonds
Beta turns, random coils also common in loops
Spatial folding of polypeptide chain in properly arranged, repetitive structures. Stabilizing forces - h bonding for alpha helix, beta sheet
ALPHA HELIX, BETA SHEET, BETA TURNS, RANDOM COIL
What defines tertiary structure, and what forces stabilize it? What 2 types?
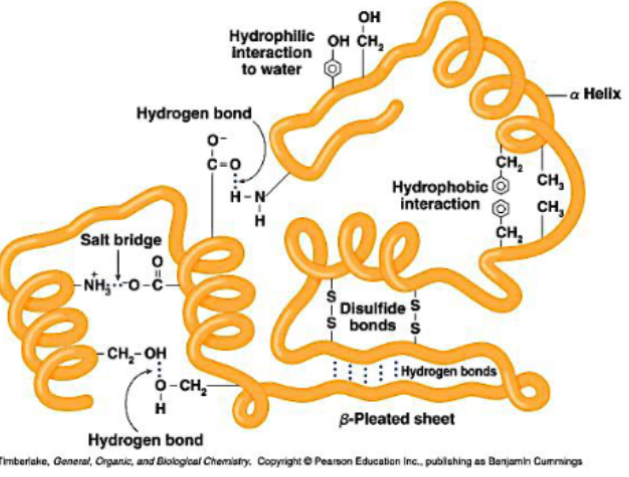
The full 3D folding of a single polypeptide chain, bringing distant amino acids into proximity.
3D arrangement of atoms in molecule - “conformational”
Stabilised by:
Non-covalent interactions: hydrophobic interactions, hydrogen bonds, ionic (electrostatic) interactions, Van der Waals
Covalent disulfide bonds between cysteine residues
Interaction with water and hydration shell also helps stability
Proteins can be fibrous, globular, or (minor) membrane proteins depending on folding.
What is quaternary structure in proteins?
It describes the assembly of two or more polypeptide (tertiary-structured) subunits into a larger complex. Subunits have own tertiary structure. Subunits may work independently or cooperatively. Dissociation of subunit results in loss of function The same non-covalent interactions (and sometimes covalent ones) stabilize quaternary structure.
Name some major classes or roles of proteins and give examples.
Enzymes: catalyse chemical reactions
Transport proteins: e.g. hemoglobin, membrane channels
Structural proteins: e.g. keratin, collagen, elastin
Receptors / signal proteins: e.g. insulin receptor
Hormones: small protein or peptide signals
Antibodies / immunoglobulins: defense proteins
Vaccines: often use protein antigens
Membrane proteins: integral or peripheral
For example: collagen gives structure in connective tissues; elastin gives elasticity to tissues; spider silk is a natural polymeric structural protein.
What are the main characteristics of enzymes?
Catalytic power: can accelerate reactions enormously (often many orders of magnitude). Define as ratio or enzymes catalysed ROR to uncatalysed rate. As much as 10²^6. In dilute as solutions under mild conditions of temp and PH
Specificity: highly selective for particular substrates
Regulation: enzyme activity is tightly controlled (pH, allosteric, inhibitors, etc.) appropriate for cellular requirements
How are enzymes classified into major groups?
Oxidoreductases — catalyse redox (oxidation-reduction) reactions
Transferases — transfer functional groups between molecules
Hydrolases — catalyse hydrolysis reactions
Lyases — break bonds without water or add to double bonds
Isomerases — rearrange atoms within a molecule
Ligases — join two molecules using energy (often from ATP)
Translocases — move ions or molecules across membranes
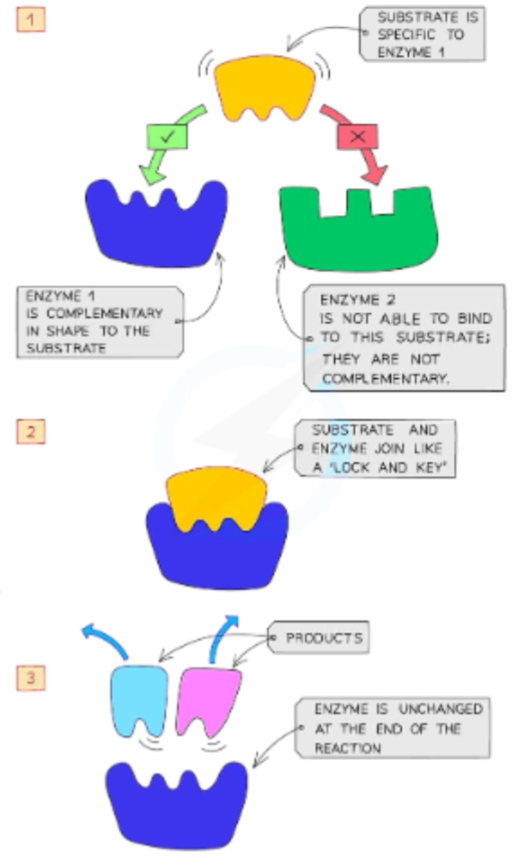
What is an active site, and how do enzymes bind substrates?
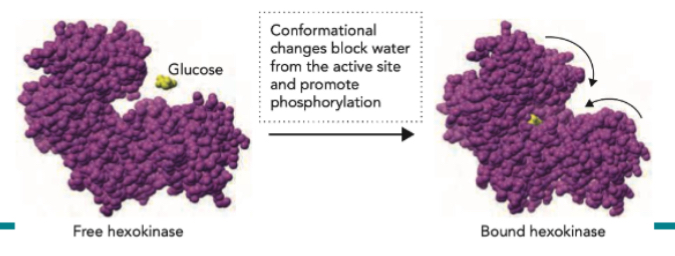
The active site is a pocket or cleft on the enzyme where substrate binds and the chemical reaction takes place. Contain reactive functional group for substrate to bind. Binding is mediated by non-covalent interactions (hydrogen bonding, ionic interactions, van der Waals). When substrate binds, the enzyme may change shape (induced fit) to enhance catalysis.
What is a catalytic triad in enzyme active sites?
It’s a set of three amino acid residues (often in hydrolases) arranged to mediate catalysis via a charge relay network: a nucleophile (commonly Ser, Cys, occasionally Thr or selenocysteine), a base, and an acid. The three act cooperatively in the catalytic mechanism.
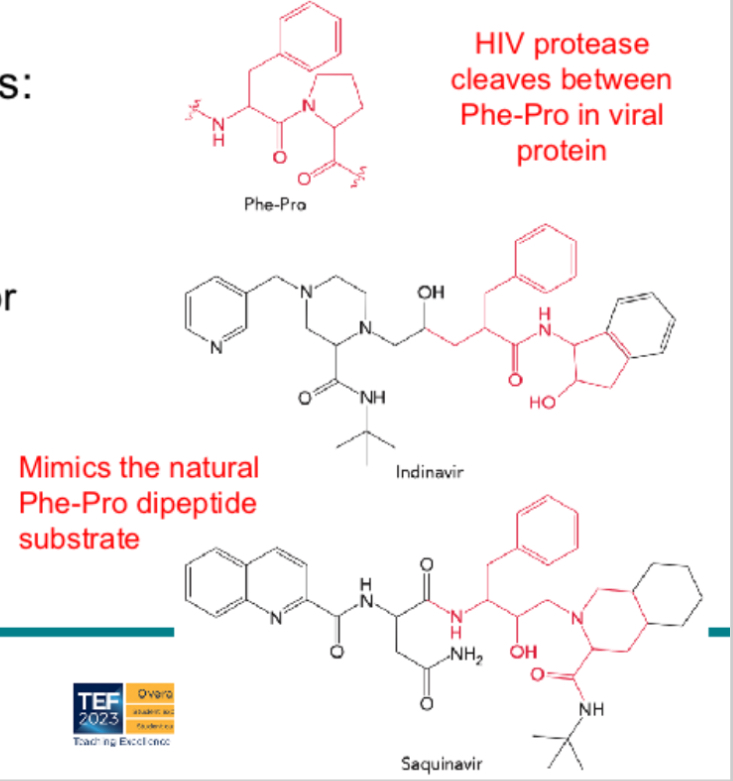
What is a competitive inhibitor, and how is it relevant in pharmacology?
A competitive inhibitor binds to the active site of the enzyme (competes with substrate) and reduces activity. In drugs, structure‑based design can produce inhibitors (e.g. HIV protease inhibitors, sulfanilamide) that mimic substrate or transition state and block the enzyme.
Sulfanilamide is an ab chemotherapeutic agent used topically for vaginal yeast infections