M5L1 - Protein Processing and Trafficking RER
1/26
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
27 Terms
Dynamics of the ER
Membranes of the ER constantly changes/undergoes
Shape/Structure
fisson/fusion
migration to new cell locations
RER (4) vs smooth ER (5)
Rough:
Ribosomal complexes associate with it
Site of co-translational transport
Site of protein modification
Formation of vesicles transporting proteins to GA
Smooth:
No ribosomes
Site of fatty acid synthesis
Site of phospholipid synthesis
Carbohydrate metabolism occurs here
Calcium is sequestered/collected here to regulate [ca]
![<p>Rough:</p><ul><li><p>Ribosomal complexes associate with it</p></li><li><p>Site of co-translational transport </p></li><li><p>Site of protein modification </p></li><li><p>Formation of vesicles transporting proteins to GA </p></li></ul><p></p><p>Smooth:</p><ul><li><p>No ribosomes</p></li><li><p>Site of fatty acid synthesis</p></li><li><p>Site of phospholipid synthesis </p></li><li><p>Carbohydrate metabolism occurs here </p></li><li><p>Calcium is sequestered/collected here to regulate [ca]</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/5e0cdcf8-ff37-4645-a057-40ee9d86003c.png)
Post-Translational Modifications (PTMs) in ER (4)
Occur along the length of entire protein if targeted to ER lumen
Occur along luminal portion if embedded in ER membrane
Types:
Glycosylation: Covalent addition of polysaccharides
Disulfide bond formation
Protein folding
Proteolytic cleavage
Protein Glycosylation
Addition of polysaccharide (sugar group) to protein
Common in
proteins to be secreted from the cell
proteins embedded in cell membrane
proteins which mediate cell interactions with extracellular matrix (space between outside of cells)
proteins that mediate receptor-ligand recognition
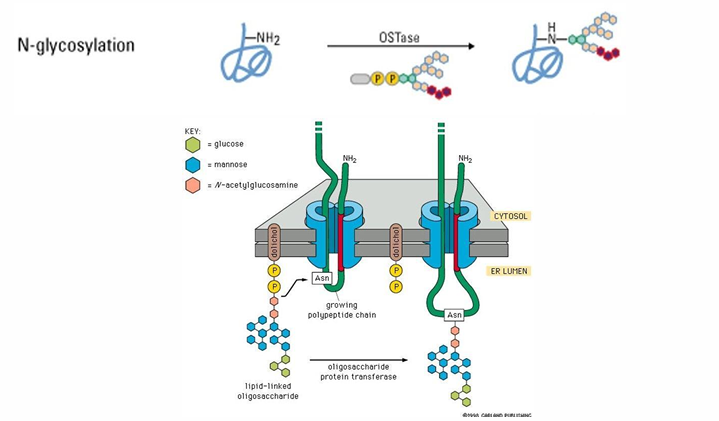
N-linked glycosylation in ER
Most common form of glycosylation
Addition of a polysaccharide to the NH2 of asparagine’s R-group
This portion of the proteins remains on the luminal side
The modification appears on the exterior surface of the protein
Disulphide bond formation in ER
Covalent linkages between the (-SH) groups of two cysteine amino acids
They provide added stability
Forms tertiary/quarternary structures of proteins
Can occur
Within single protein (intramolecular bond)
Between proteins (intermolecular bond)
Common in proteins
secreted from the cell
located on the outside surface of cell membrane
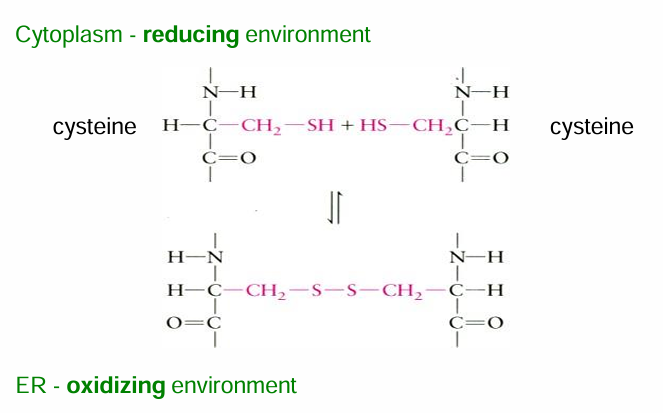
ER lumen has an oxidative environment favouring the formation of this bond (it’s an oxidation rxn)
Cytosol has a reducing env favouring the rev rxn
RNAse A: Disulphide Bridge Protein Example
has 4 disulphide bridges
Secreted into intestine to aid in digestion of RNA by cleaving it into smaller pieces
The bonds help maintain the structure in the acidic env of small intestine
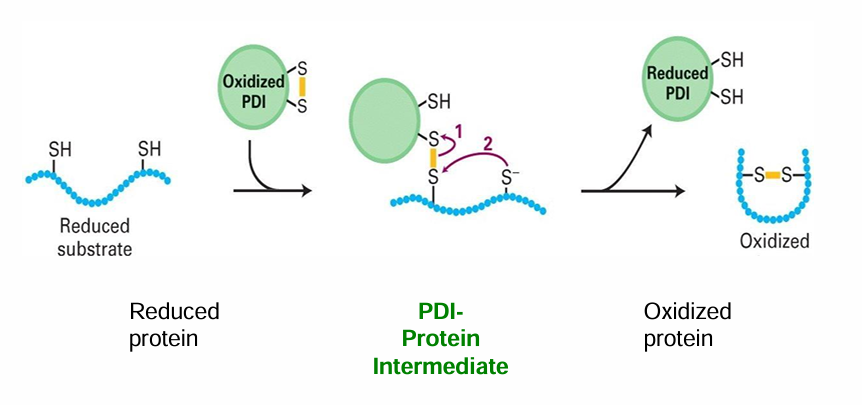
Protein Disulphide Isomerase (PDI)
ER protein that promotes oxidation, thus promoting disulphide bridge formation
PDI can also correct inappropriate disulfide bridge formations
Forms an intermediate with 2 cysteine residues to accelerate rate of rxn
Steps:
Oxidized PDI contains a disulfide bridge
PDI forms an intermediate with one cysteine residue
This facilitates the formation of an intramolecular cysteine bond
PDI is spontaneously converted back into the oxidized form bc of the oxidizing env of the ER luman
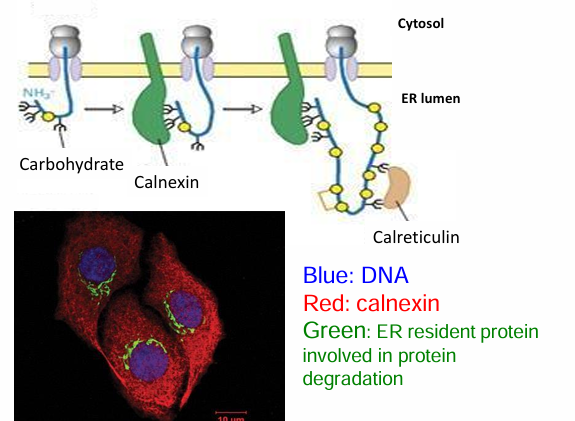
Protein Folding in the ER (Lectins)
Lectins assist protein folding by recognizing N-linked polysaccharides on them (from glycosylation)
They function similarily to molecular chaperones
Lectins types:
Calnexin: Found throughout ER membrane
Calreticulin
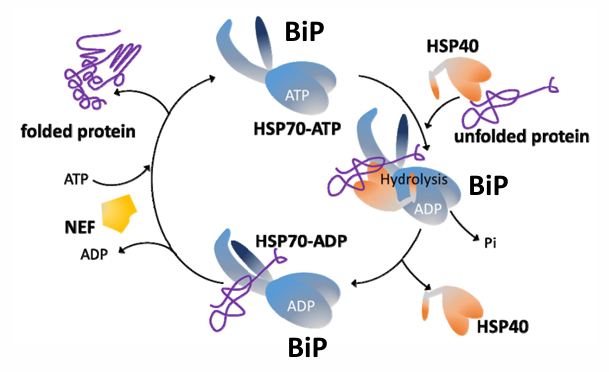
BiP: HSP70
Is an ER protein
The roles of BiP depend on its ability to recognize and bind to unfolded proteins
It binds to proteins as they appear in the ER through co-translational transport
BiP and it’s co-chaperones (Hsp40 and NEF) drive efficient ER protein folding
BiP initiates a process called the unfolded protein response in the ER
Proteolytic Cleavage in the ER
Cleavage of peptide backbone of protein
Ex. N-terminal signal cleavage by signal peptidase
Ex. Proinsulin
Has N-terminal sequence removed by signal peptidase
Required to form the final insulin protein in pancreatic cells
Has disulfide bridges providing extra stability
Proinsulin
Has N-terminal sequence removed by signal peptidase
Required to form the final insulin protein in pancreatic cells
Has disulfide bridges providing extra stability
Is the target of 3 other peptidases during transport in pancreatic cells through secretory vesicles
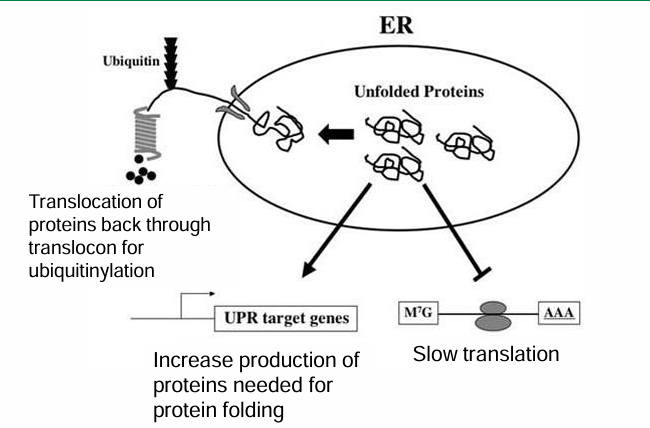
Unfolded Protein Response (UPR)
The RER can be flooded with unfolded proteins resulting from overproduction, delay in processing, exposure to toxins, denaturing, or lack of nutrients
This causes a risk of aggregation and cell death
The UPR is a response to this situation
First response: Try to restore normal function by slowing down new protein translation or removing unfolded proteins through ubiquitinylation
Second Step: Increase production of chaperone proteins assisting in folding
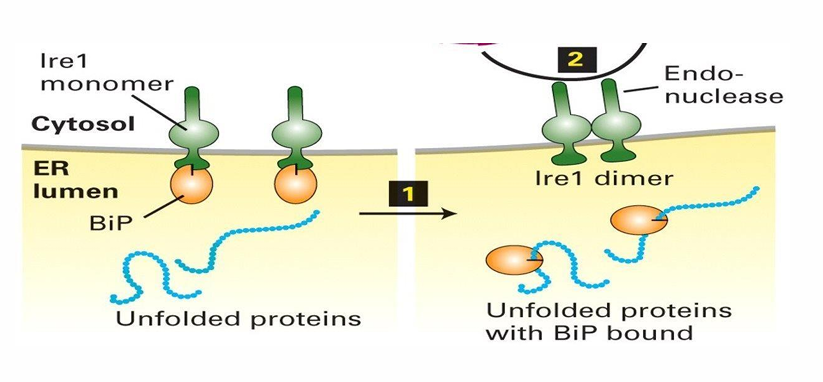
BiP and Ire1 in UPR
BiP serves as chaperone to assist in folding and prevent aggregation
BiP is part of the system leading to the production of more protein-folding regulators
BiP and lre1 associated
BiP is inhibited and lre1 is inactive
Increase in unfolded proteins causes BiP and lre1 to disassociate
BiP has higher affinity for hydrophobic pathes on unfolded proteins
lre1 forms homodimers in the ER which serve as activated endonucleases
Endonucleases make internal cuts in nucleic acids like mRNAs
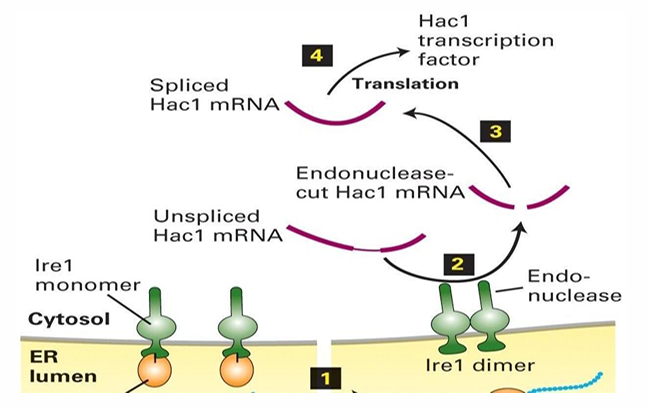
Hac1 Expression during UPR
lre1 endonuclease targets the mRNA for Hac1
Unspliced Hac1 mRNA has an internal sequence inhibiting translation by ribosome
lre1 splices Hac1 mRNA to allow for the synthesis of Hac1 protein
It serves as a transcription factor
It’s transported to the nucleus to activate the transcription of several genes
This includes BiP, lectins, PDI and signal peptidases
Protein Trafficking Away / To ER
Anterograde Transport: Proteins move from ER to the Cell membrane (away from ER)
Retrograde Transport: Proteins brought back to ER
Techniques for Studying Vesicular Transport (from ER to out of cell)
Pulse-chase labeling and visualization using immuno-TEM
Fluorescent microscopy of GFP-labelled proteins
Both in mammalian cells
Genetiv mutations disrupting transport in yeast cells allowed for identification of the proteins required
Pulse-Chase Labeling: Acinar cells
Acinar cells: exocrine cells of the pancreas
they produce and transport enxymes secreted into digestive system
Tagging the secreted enzymes can show us where they go after leaving the ER
Pulse-chase:
Tagging proteins for a brief amount of time so only some are labelled
Acinar cells were incubated in a medium with radioactive methionine
This is a ‘pulse’ of labelling which lasted 3 mins
Some cells are removed from the medium, washed, and transferred into a medium with non-radioactive amino acids
This ‘chase’ can vary in length so proteins can be visualized at different stages in their journey from RER to secretion
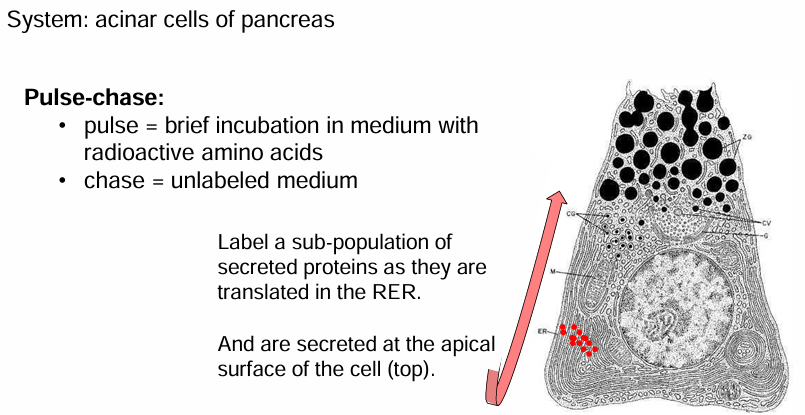
Pulse-Chase Labeling Different Chase Lengths
Time Point 1:
Cells grown in radioactive medium for 3 mins
They’re fixed immediately with no ‘chase’
They’re visualized using electron microscopy
They’re found in the ER bc they haven’t moved
Time Point 2:
Cells grown in radioactive medium for 3 mins
Allowed to be in unlabelled medium for 17 mins (protein transport occurs)
They’re then fixed and visualized
They’re found in the GA
Time Point 3:
Cells grown in radioactive medium for 3 mins
Allowed to be in unlabeled medium for 117 mins
They’re then fixed and visualized
The proteins are found in secretory vesicles
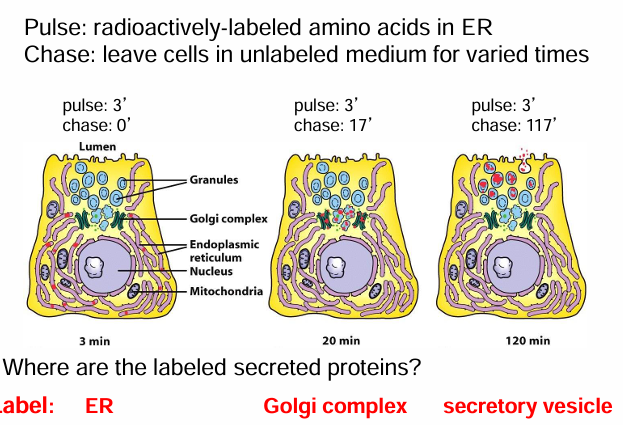
Pulse-Chase Graph: Location of Labeled Proteins Throughout Experiment
y axis: % of autoradiographic grains on TEM image (due to radioactively-labelled proteins)
x axis: time (length of chase) from 0-120 mins
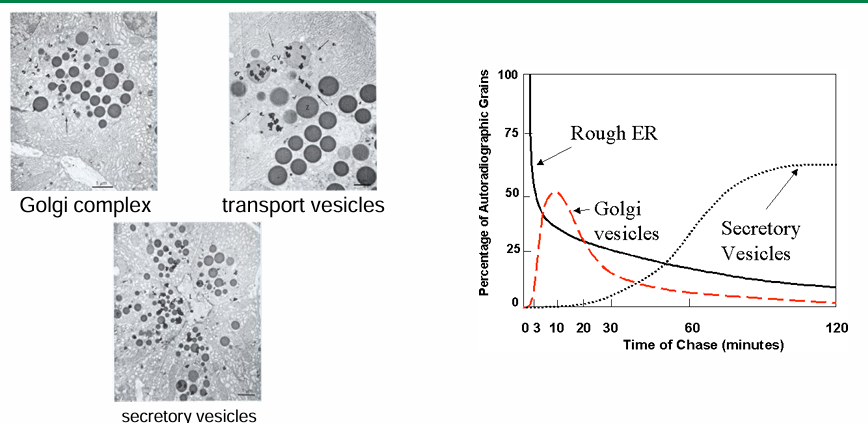
GFP Tagging of VSV (Vesicular Stomatitis Virus)
Carries a gene coding for an envelope protein (G protein)
Viral proteins can be synthesized and embedded in host membrane
It becomes part of the viral envelope surrounding the virus then
A mutant VSV-G protein can be tagged
At permissive temp (32C) the protein variant can fold and will be found in cell membrane
At restrictive temp (40C) the protein variant denatures and is retained in ER by UPR quality control
Pulse-Chase GFP Tagging of VSV (Vesicular Stomatitis Virus)
Infect mammalian cells with viruses carrying VSV-G:GFP gene with enough time to induce protein synthesis
Disable the virus and culture the mammalian cells at 40C
VSV-G:GFP is retained in ER
Lower temp to 32C
VSV-G:GFP will fold and be transported out
Tracked movement
0 mins: temp not shifted for protein accumulates in ER
40 mins after move to 32C: Fluorescent protein moved to GA
180 mins after move to 32C: fluorescent protein moved to membrane
Graph:
Summarizes location of VSV-G:GFP protein
y-axis: amount of fluorescence in cell in arbitrary units
x-axis: length of chase from 0-600 mins
Fluorescence dec overtime as fluorophores lose their fluoresence as they move from ER to GA to membrane
Saccharomyces cerevisiae (yeast) model in determining steps in protein transport
It was for protein transport to cell membrane from ER
The yeast S. cerevisiae metabolizes sucrose by hydrolyzing it into glucose and fructose
This is done by protein invertase created by another cell
Defects in protein transport pathways can be identified by following the secretion of invertase
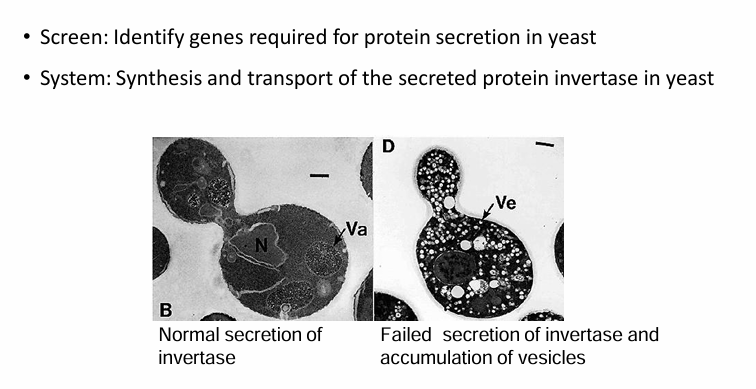
Invertase Secretion:
Generated random mutations in yeast genome
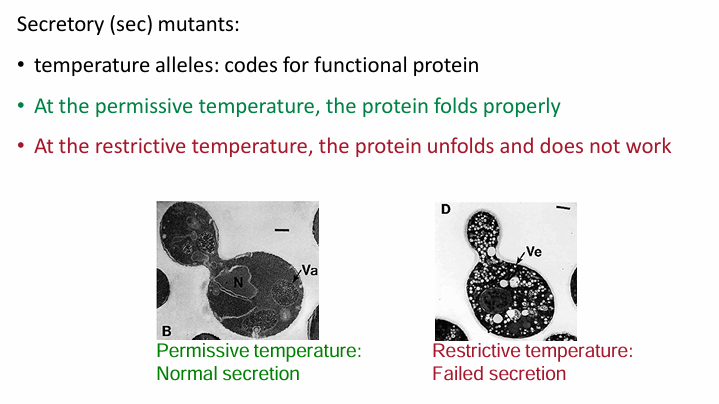
They looked for temp-sensitive mutations than failed to secrete invertase in restrictive temps
The mutations code for proteins folding normally at permissive temp but not in restrictive temp
A restrictive temperature shift will reveal defect in transport (invertase will accumulate in vesicles)
Each gene mutation was called a sec mutant (secretory mutant)
They were named by number given the place of mutation
Ex. Sec61 (translocon protein)
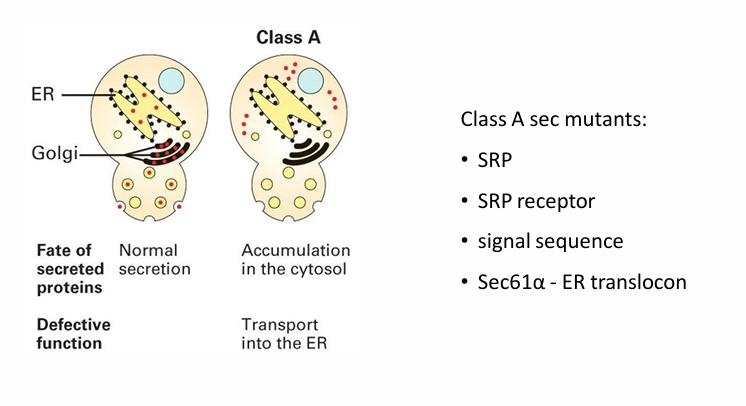
Class A sec mutants (yeast experiment)
Invertase accumulated in different locations based on which step was defective
Different classes were identified based on the location of accumulation
Class A showed accumulation in cytosol
Normally it would be in the ER, GA, vasicles, and outside the cell
It suggested the mutation prevents the first step of co-translational transport
This would be linked to any component of ER translocon
SRP protein
SRP receptor
signal sequence
Sec61-alpha (ER translocon)
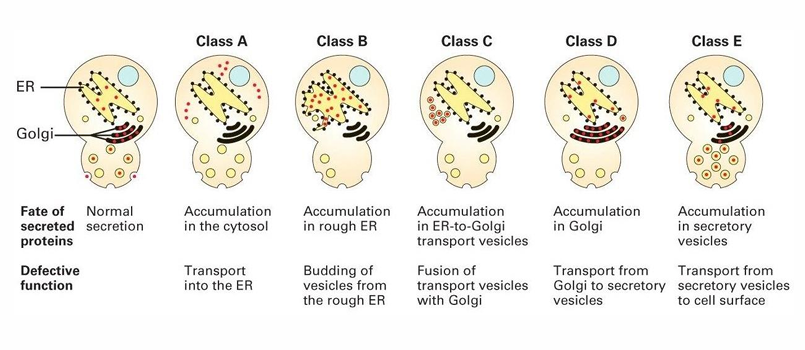
The 5 Classes of sec mutants (yeast invertase study)
Class A: Accumulates in cytosol
Class B: Accumulates in ER
Defect in vesicle transport out of ER
Class C: Accumulates in ER to Golgi transport vesicles
Class D: Accumulates in in GA
Defect in forming vesicles to leave GA
Class E: Accumulates in secretory vesicles to secrete out of cell
Double Mutants in Yeast Experiment to Determine Protein Transport
Cells carrying 2 mutations from a different class each
This could demonstrate the step-wise progression of transport
Ex. Cell with Class A and B mutations would only show a Class A phenotype
Therefore, Class A proteins are needed before Class B ones
Upstream mutations mask the appearance of downstream phenotypes
A collection of double-mutants confirmed the pathway from RER to GA to vesicles