Biochem Lect 32: ETC, Oxidative Phos
1/70
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
71 Terms
purpose of electron transport
create proton gradient using electron movement
purpose of oxidative phosphorylation
use proton gradient to make ATP
Enzymes of each Complex
Complex I = _____________
Complex II = _____________
Complex III = _____________
Complex IV = _____________
Complex V = _____________
Enzymes of each complex
Complex I = NADH dehydrogenase
Complex II = Succinate CoQ Reductase
Complex III = UQ-Cytochrome C Reductase
Complex IV = Cytochrome C Oxidase
Complex V = ATP Synthase
Order of Complexes
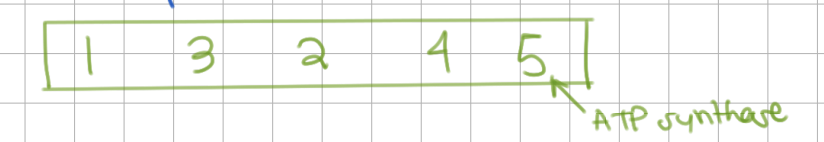
Which complexes contain Fe-S clusters or electron wires?
Complex I = Fe-S clusters and electron wire
Complex II = Fe-S clusters only
Complex III = 1 Fe-S cluster (Rieske)
Complex IV = None
Complex V = None
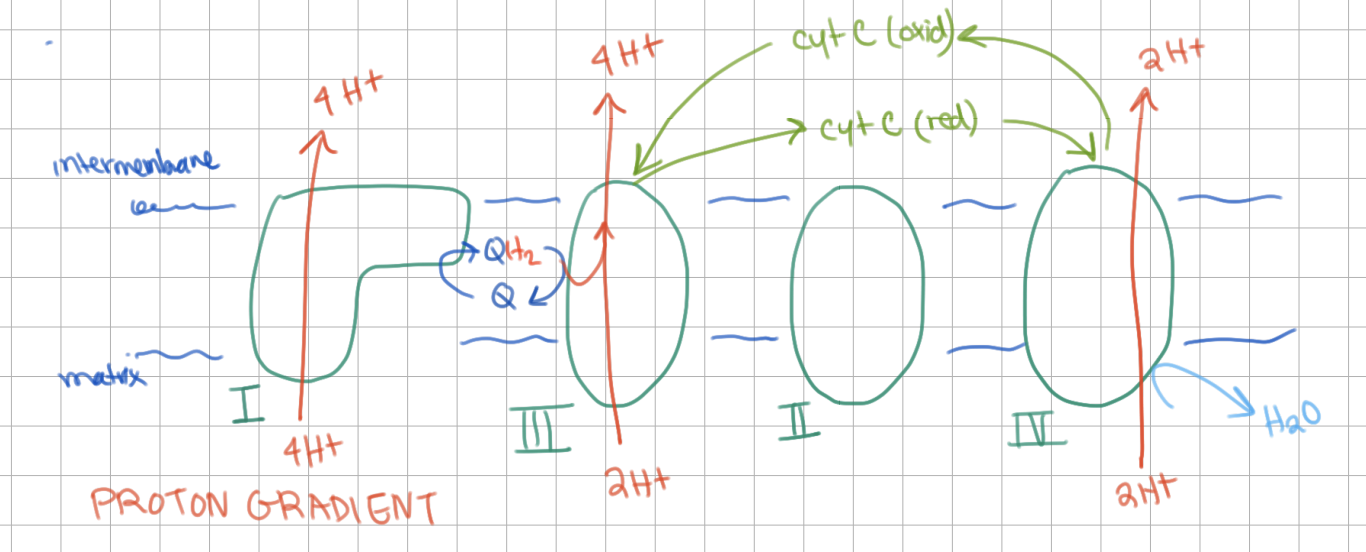
How many protons does each complex pump?
Complex I = 4
Complex II = 0
Complex III = 4
Complex IV = 2
Complex I = ?
NADH dehydrogenase
Complex I Structure
includes a large matrix arm on matrix side
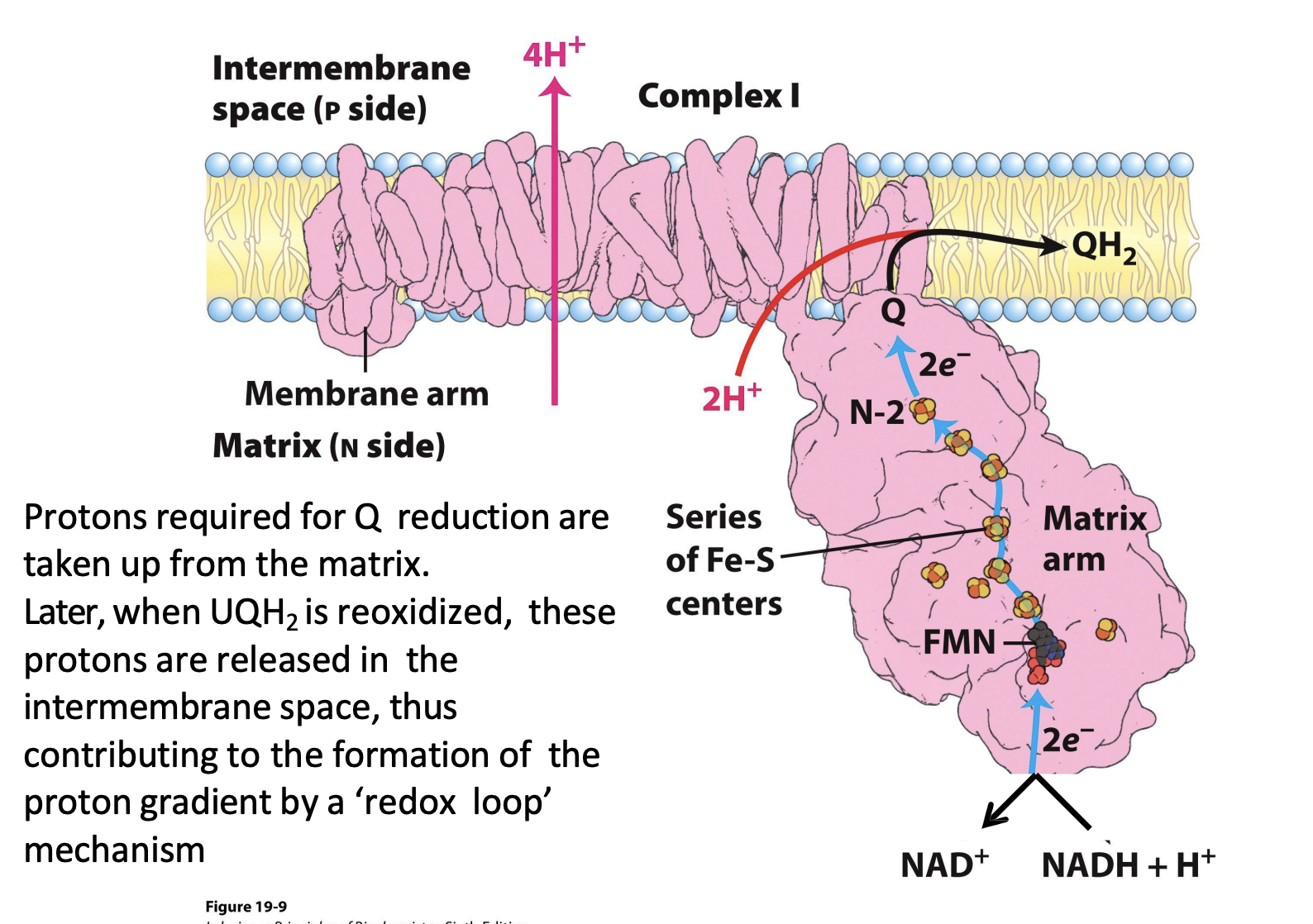
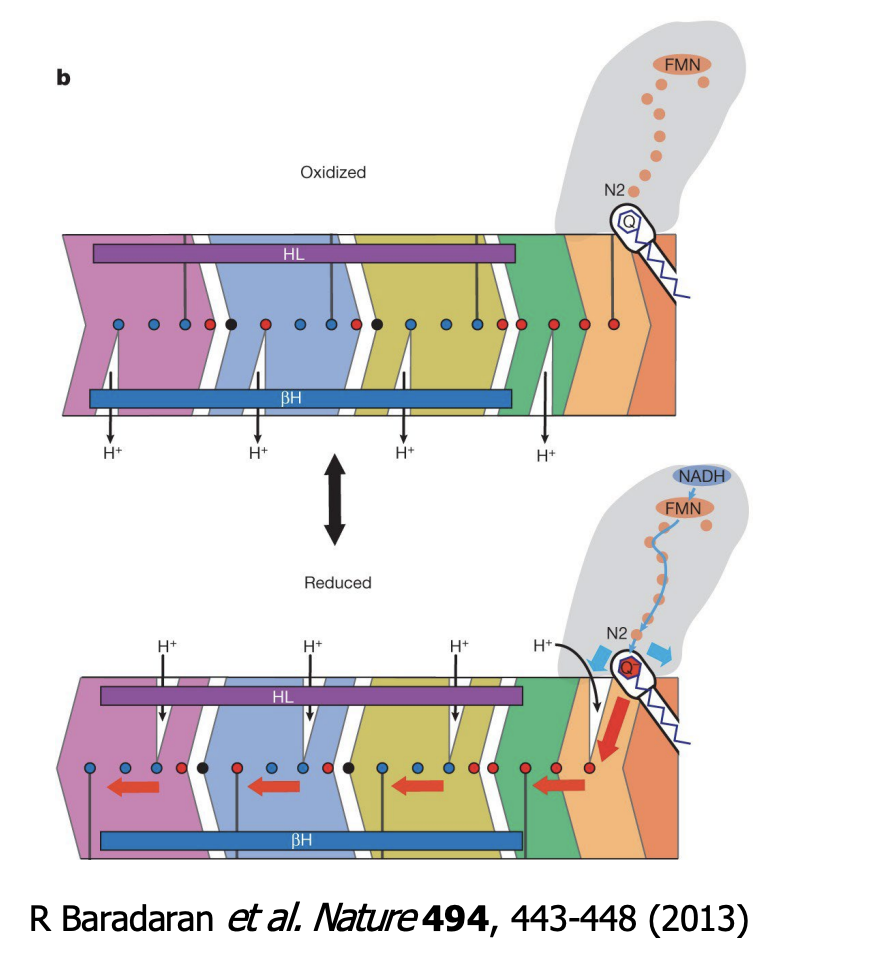
___________ (on the NuoL subunit) on Complex I helps drive _________ that couples electron transfer to proton pumping
A long alpha helix (on the NuoL subunit) on Complex I helps drive conformational changes that couples electron transfer to proton pumping
Ubiquinone = ________, _____-soluble electron carrier
Also known as: _______ (____), ____, ____
Reduced form: ____
Ubiquinone = coenzyme, lipid-soluble electron carrier
Also known as: Coenzyme Q (CoQ), UQ, Q
Reduced form: QH2
Complex I Mechanism
1) ______ gets oxidized by matrix arm → sends ____ e-
2) ///
3) ///
Complex I Mechanism
1) NADH gets oxidized by matrix arm → sends 2 e-
2) ///
3) ///
Complex I Mechanism
1) NADH gets oxidized by matrix arm → sends 2 e-
2) ____ gets 1 electron at a time from NADH to send through ________ within matrix arm
3) ///
Complex I Mechanism
1) NADH gets oxidized by matrix arm → sends 2 e-
2) FMN gets 1 electron at a time from NADH to send through electron wire within matrix arm
3) ///
Complex I Mechanism
1) NADH gets oxidized by matrix arm → sends 2 e-
2) FMN gets 1 electron at a time from NADH to send through electron wire within matrix arm
3) ___ gets reduced (gains ___ e-) and transfers electrons to Complex ___
Complex I Mechanism
1) NADH gets oxidized by matrix arm → sends 2 e-
2) FMN gets 1 electron at a time from NADH to send through electron wire within matrix arm
3) Q gets reduced (gains 2 e-) and transfers electrons to Complex III
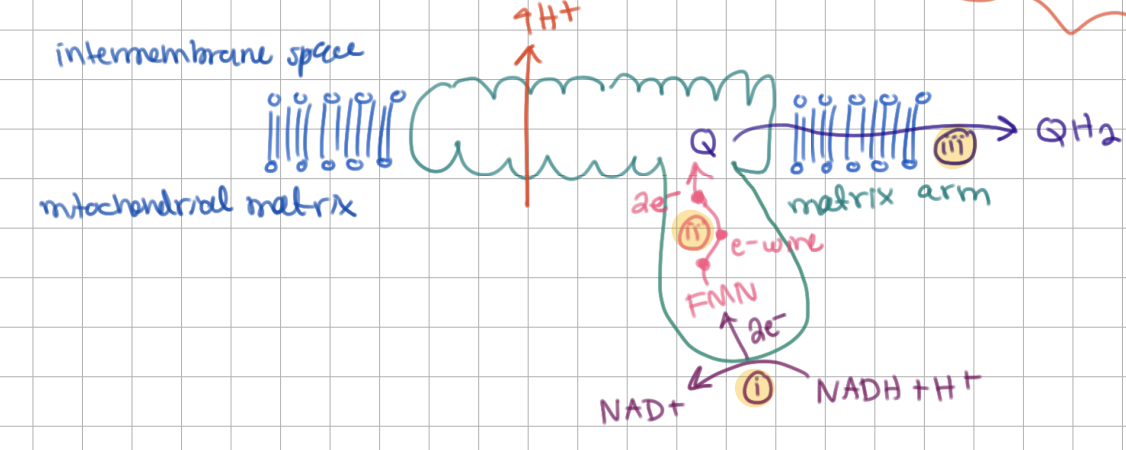
How many H+ are pumped across membrane in Complex I (for every 2e-)?
4 H+
Q gets reduced to QH2 and later QH2 will get re-oxidized. What is the phenomenon called?
redox loop
Complex II = ?
Succinate-CoQ Reductase
Alternate names for Complex II
succinate dehydrogenase (from TCA cycle)
flavoprotein 2 (FP2) → contains FAD
Complex II Structure
___ type cytochrome
contains ___ subunits
___ Fe-S clusters (…)
contains ___
contains non-covalently bound _____
Complex II Structure
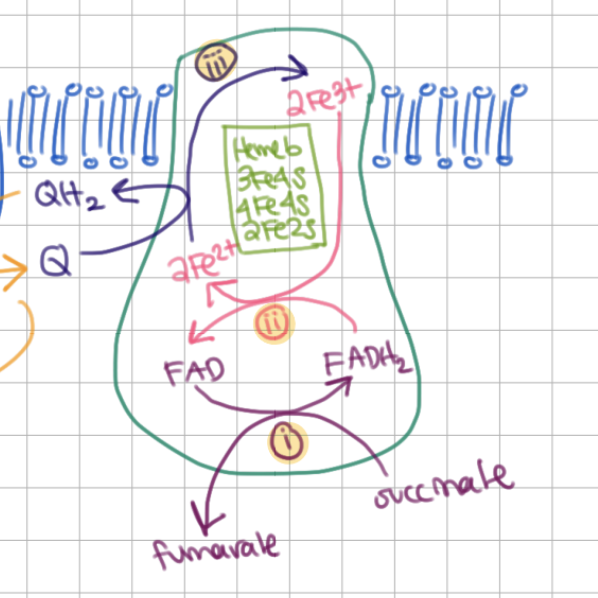
b type cytochrome
contains 4 subunits
3 Fe-S clusters (3Fe4S, 4Fe4S, 2Fe2S)
contains FAD ← flavoprotein 2
contains non-covalently bound heme b (Fe protophyrin IX)
Can heme participate in redox? Is it involved in the complex II reaction?
Yes, it can transfer 1 electron.
However in this reaction, it is not involved.
How many protons are pumped across complex II?
0
Does complex II contain an electron wire?
no
Complex II Mechanism
1) ________ oxidizes to ________ ←→ ____ reduces to ____
2) ///
3) ///
Complex II Mechanism
1) succinate oxidizes to fumarate ←→ FAD reduces to FADH2
2) ///
3) ///
Complex II Mechanism
1) succinate oxidizes to fumarate ←→ FAD reduces to FADH2
2) FADH2 oxidizes to FAD ←→ 2___ reduce to 2____
3) ///
Complex II Mechanism
1) succinate oxidizes to fumarate ←→ FAD reduces to FADH2
2) FADH2 oxidizes to FAD ←→ 2Fe3+ reduce to 2Fe2+
3) ///
Complex II Mechanism
1) succinate oxidizes to fumarate ←→ FAD reduces to FADH2
2) FADH2 oxidizes to FAD ←→ 2Fe3+ reduce to 2Fe2+
3) 2Fe2+ oxidize to 2Fe3+ ←→ ____ reduces to ____
Complex II Mechanism
1) succinate oxidizes to fumarate <=> FAD reduces to FADH2
2) FADH2 oxidizes to FAD ←→ 2Fe3+ reduce to 2Fe2+
3) 2Fe2+ oxidize to 2Fe3+ ←→ Q reduces to QH2
As complex II reduces Q, complex ___ oxidizes QH2
As complex II reduces Q, complex III oxidizes QH2
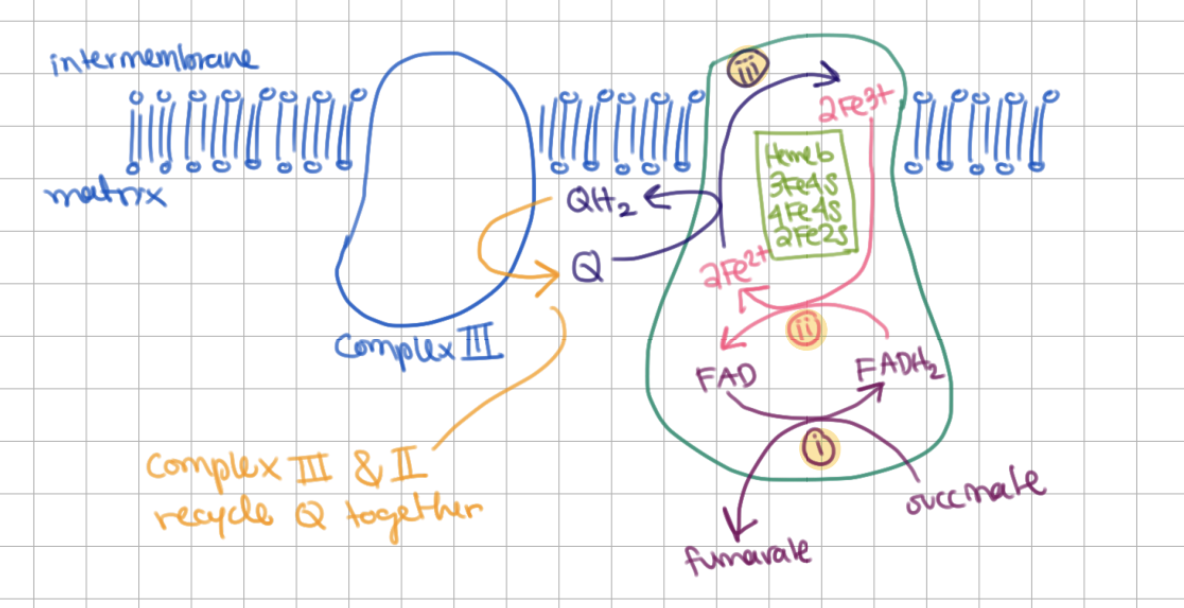
electrons flow in what order for the follwing:
FAD, heme B, Fe-S clusters

Succinate creates more/less ATP than NADH (and why?)
NADH gives electrons to which complexes?
Succinate gives electrons to which complexes?
less protons = more/less ATP
Succinate creates less ATP than NADH
NADH gives electrons to complex I (4 H+) → III → IV → V
Succinate gives electrons to complex II (0 H+) → III → IV → V
less protons = less ATP
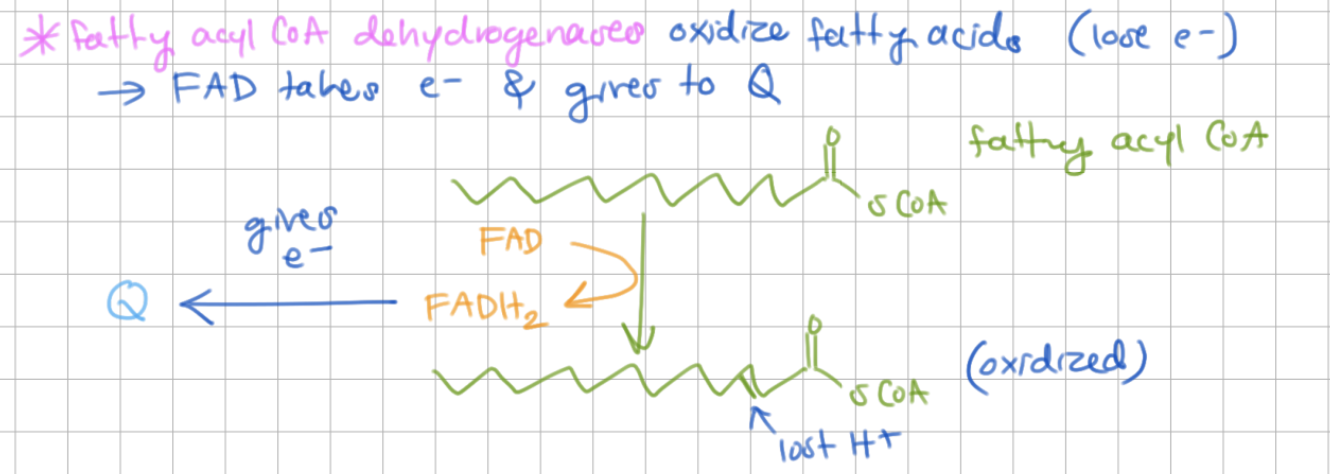
_______________ oxidize fatty acyl CoAs and give electrons to Q.
FAD/NAD takes electrons.
Fatty acyl CoA dehydrogenase oxidize fatty acyl CoAs and give electrons to Q.
FAD takes electrons.
Complex III = ?
UQ-Cytochrome C Reductase
Complex III Structure
contains ___ cytochrome which contains hemes ___ and ___
Complex III Structure
contains b cytochrome which contains hemes bL and bH
Q Cycle Phase I
QH2 binds to ___ site and gets reduced to ___ → releases ___H+
///
///
Q Cycle Phase I
QH2 binds to Qp site and gets reduced to Q → releases 2H+
///
///
Q Cycle Phase I
QH2 binds to Qp site and gets reduced to Q → releases 2H+
1st electron gets transferred to ___ → ___ → ___
///
Q Cycle Phase I
QH2 binds to Qp site and gets reduced to Q → releases 2H+
1st electron gets transferred to Fe-S (in Rieske subunit) → cyt c1 → cyt c
///
Q Cycle Phase I
QH2 binds to Qp site and gets reduced to Q → releases 2H+
1st electron gets transferred to Fe-S (in Rieske subunit) → cyt c1 → cyt c
2nd electron gets transferred to ___ → ___ → reduces Q → ___ in ___ site
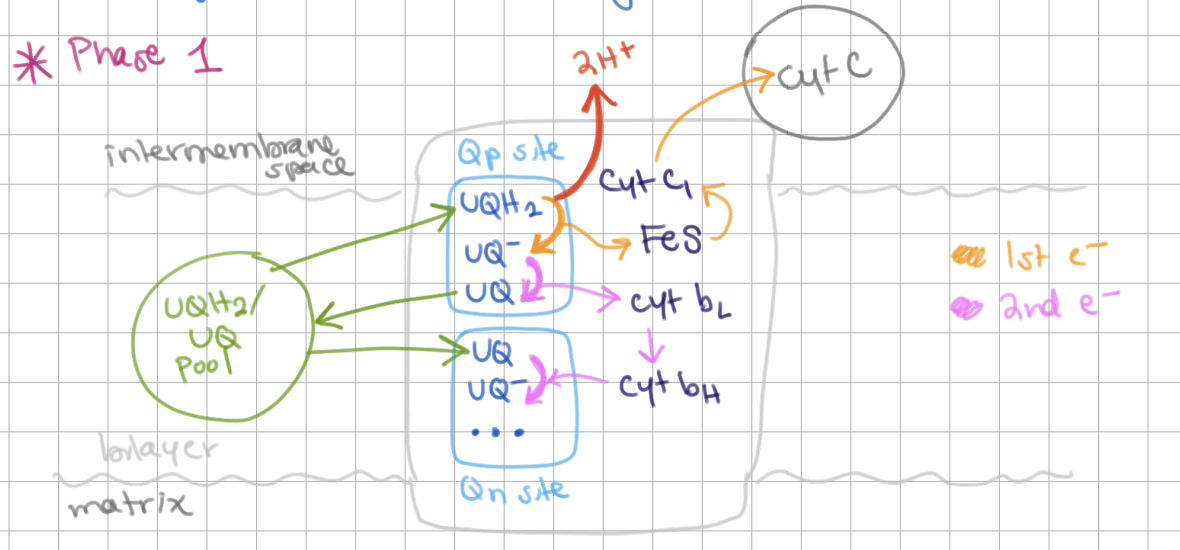
Q Cycle Phase I
QH2 binds to Qp site and gets reduced to Q → releases 2H+
1st electron gets transferred to Fe-S (in Rieske subunit) → cyt c1 → cyt c
2nd electron gets transferred to cyt bL → cyt bH → reduces Q → Q- in Qn site
Q Cycle Phase II
A second QH2 binds to Qp site and gets reduced to Q → releases 2H+
At Qn site, ___ is able to reduce to ___ (requires ___H+) → ___ is released into __________.
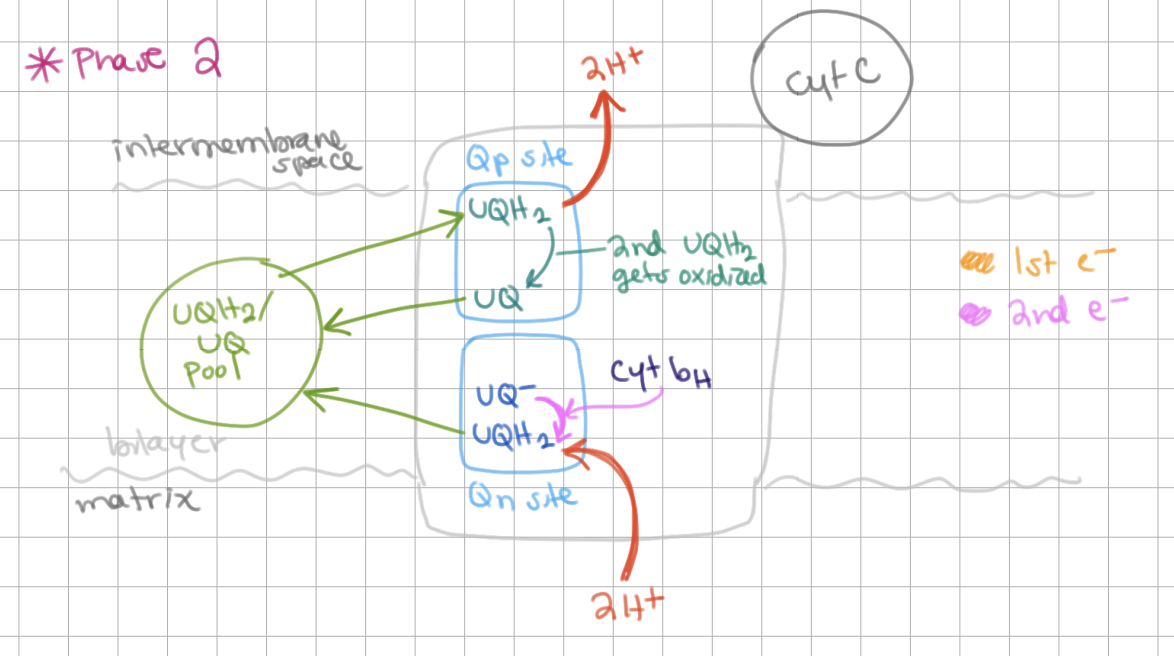
Q Cycle Phase II
A second QH2 binds to Qp site and gets reduced to Q → releases 2H+
At Qn site, Q- is able to reduce to QH2 (requires 2H+) → QH2 is released into lipid bilayer.
cytochrome C is an electron tranferer (transfers ___ e-)
it is ______ soluble, so it moves in the ______
(QH2 is ______ soluble, so it moves in the ______)
cytochrome C is an electron tranferer (transfers 1 e-)
it is water soluble, so it moves in the intermembrane space
(QH2 is lipid soluble, so it moves in the bilayer)
cytochrome C passes electrons from complex ___ to ___
cytochrome C passes electrons from complex III to IV

cytochrome C structure
total ___ sulfurs
____ at center, covalently linked with ___ S
___ S coordinates ___
cytochrome C structure
total 3 sulfurs
heme at center, covalently linked with 2 S
3rd S coordinates Fe 2+ (in heme)
Complex IV = ?
Cytochrome C Oxidase (COX)
COX oxidizes ________, reduces ________ (___ electrons)
→ ___ H2O released
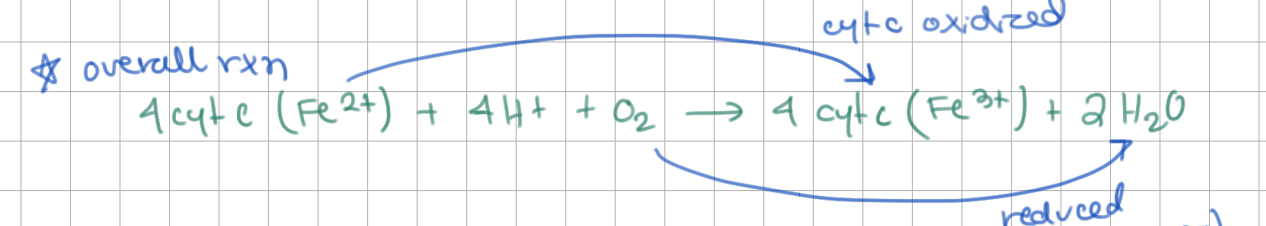
COX oxidizes cytochrome C, reduces O2 (4 electrons)
→ 2 H2O released
_________ is the terminal oxidoreductase, ___ is the final acceptor of electrons.
COX is the terminal oxidoreductase, O2 is the final acceptor of electrons.
How is 2 H2O created in Complex IV?
4H+ reduces O2
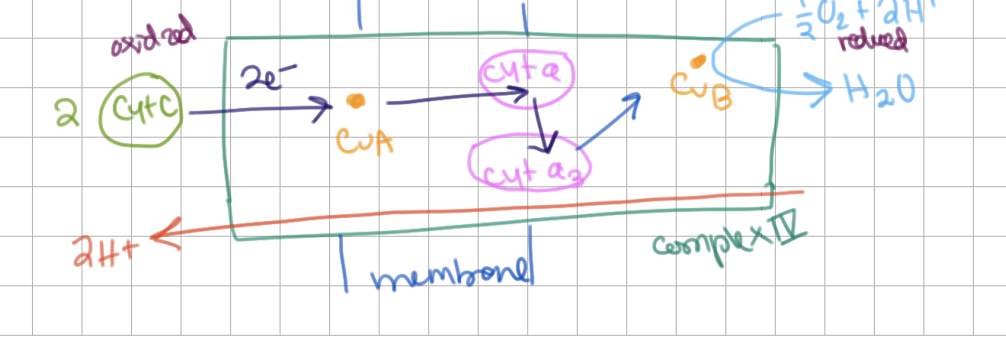
COX Structure
How many hemes and how many coppers?
2 hemes (a and a3)
2 copper sites (CuA and CuB)
COX Mechanism
(idk if i need details, view image)
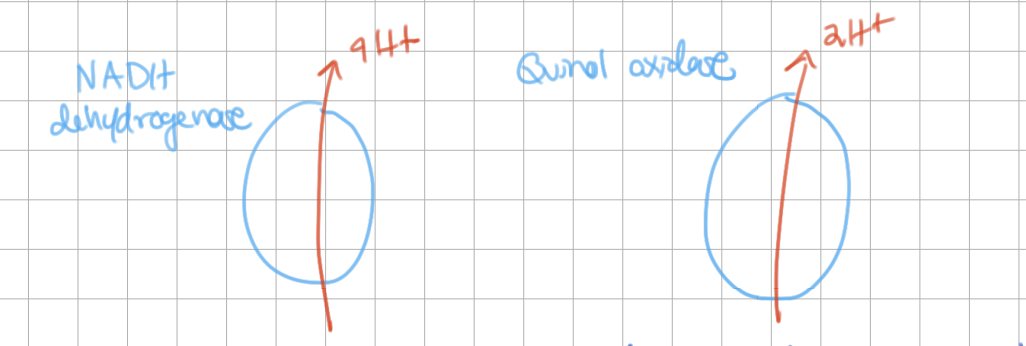
E. coli ETC (view image)
Mitchell Hypothesis = ?
proton gradient across the inner membrane drives ATP synthesis
ridiculed but right, won a Nobel prize
aka chemiosmotic hypothesis
In complexes I-IV, are H+ released into intermembrane space or matrix?
intermembrane space
electrons pass through ETC and H+ is released into intermembrane space → electric potential and proton gradient = (2 names)
electrochemical proton gradient,
proton motive force
many/few protons are present in IMS → goes back to matrix with/against concentration gradient (through _________)
many protons are present in IMS (due to ETC) → goes back to matrix with concentration gradient (through ATP synthase)
Evidence of Mitchell Hypothesis
pH increases/decreases when O2 added to respiring mitochondria
ATP synthesis stops if inner/outer membrane is disrupted
placing mitochondria in water causes swelling → ___ leak → no ATP synthesis
ATP synthesis inhibitors work by disrupting ___________
Evidence of Mitchell Hypothesis
pH decreases when O2 added to respiring mitochondria (more H+)
ATP synthesis stops if inner membrane is disrupted
placing mitochondria in water causes swelling → H+ leak → no ATP synthesis
ATP synthesis inhibitors work by disrupting proton gradient
2 types of ATP synthesis ihibitors
1) …
2) …
both types are hydrophobic/hydrophilic
1) uncouplers = equalize H+ concentration
2) ionophores = poke pores into membrane → dissipate H+ gradient
both types are hydrophobic
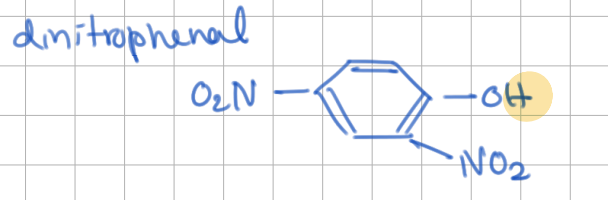
Uncouplers
uncouples _____________ and _____________
hydrophobic molecules with ____________
(how do they equalize gradient?)
Uncouplers
uncouples electron transport and proton gradient
hydrophobic molecules with dissociable proton
shuttle across membrane while carrying proton → equalizes gradient
Chemiosmotic coupling
electron transfer coupled with H+ gradient
It is thermodynamically favorable for protons to move up or down gradient?
down!!

Approximate number of protons pumped per 2 electrons
succinate = ____
NADH = ____
Approximate number of protons pumped per 2 electrons
succinate = 6
NADH = 10
ATP Synthase
-
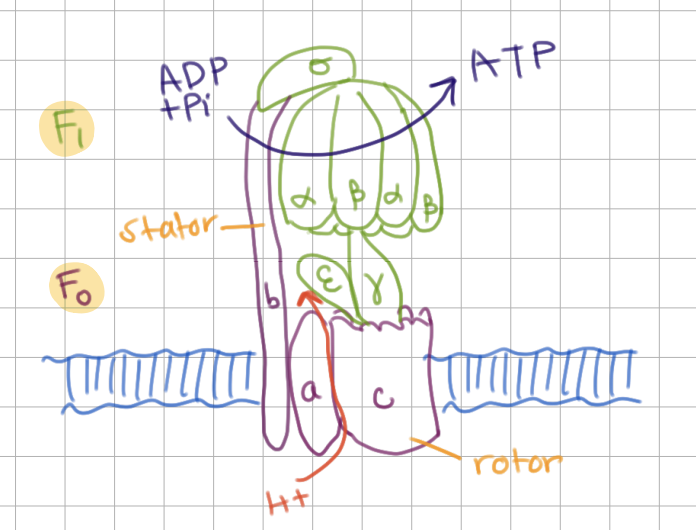
ATP Synthase has 2 main components:
F1 (subunits = ________)
F0 (subunits = ________)
ATP Synthase has 2 main components:
F1 (subunits = ɑ, β, ɣ, ε, σ)
F0 (subunits = a, b, c)
ATP Synthase subunits
____, ____ → make ATP
____ → make up top of stalk
____, ____ → middle portion (like umbrella handle)
____ → site of H+ entry (part of _____)
____ → makes up stalk (part of _____)
____ → make up channel (_____)
ATP Synthase subunits
ɑ, β, → make ATP
σ → make up top of stalk
ɣ, ε → middle portion (like umbrella handle)
a → site of H+ entry (part of stator)
b → makes up stalk (also part of stator)
c → make up channel (rotor)
ATP Synthase Mechanism
H+ flows from ___ through ___ and back to ___ which turns ________ → turns ɣ
This causes conformational changes in ___ and ___
ATP Synthesis
ATP Synthase Mechanism
H+ flows from a through c and back to a which turns rotor → turns ɣ
This causes conformational changes in ɑ and β
ATP Synthesis
F1 structure of ATP Synthase
John Walker discovered that F1 contains 3 β subunits:
contains ____ and a honhydrolyzable ___ bond
contains ____
empty
F1 structure of ATP Synthase
John Walker discovered that F1 contains 3 β subunits:
contains AMP-PNP and a honhydrolyzable β-ɣ bond
contains ADP
empty
Paul Boyer’s Binding Change Mechanism
The 3 β subunits of F1 have different conformations:
tight = contains ___ = high/low/no affinity for ADP
loose = contains ___= high/low/no affinity for ADP
open = contains ___= high/low/no affinity for ADP
Which one makes ATP? _____
Paul Boyer’s Binding Change Mechanism
The 3 β subunits of F1 have different conformations:
tight = contains AMP-PNP = high affinity for ADP
loose = contains ADP = low affinity for ADP
open = contains nothing = no affinity for ADP
Which one makes ATP? tight
Overall order of conformation switch in Paul Boyer’s mechanism
O → L → T → O …
Paul Boyer’s Binding Change Mechanism
O → L
____ presses against β subunit, kind of closing it
ATP/ADP does what?
L → T
____ rotates and presses more against β subunit, completely closing it
ATP/ADP does what?
T → O
____ no longer presses against β, relaxes
ATP/ADP does what?
Paul Boyer’s Binding Change Mechanism
O → L
ɣ presses against β subunit, kind of closing it
ADP/Pi enter
L → T
ɣ rotates and presses more against β subunit, completely closing it
ADP/Pi are forced together → ATP
T → O
ɣ no longer presses against β, relaxes
ATP leaves

In ATP Synthase, H+ enter the a subunit _____ half channel → c → a _____ half channel.
In ATP Synthase, H+ enter the a subunit inlet half channel → c → a outlet half channel.
One complete rotation of rotor produces ____ ATP molecules
3
A and C Subunit Residues
1) ____ (___) on a subunit (end of inlet half channel)
2) ____ (___) on a subunit (between half channels) moves H+ to c subunit
long side chain to pick up H+ easily
cannot be replaced with ___
3) ____ (___) on c subunit
can be replaced with ___
4) ____ (___) on a subunit (end of outlet half channel)
A and C Subunit Residues
1) Asn214 (N) on a subunit (end of inlet half channel)
2) Arg210 (R) on a subunit (between half channels) moves H+ to c subunit
long side chain to pick up H+ easily
cannot be replaced with K
3) Asp61 (D) on c subunit
can be replaced with E
4) Ser206 (S) on a subunit (end of outlet half channel)
-
**NeRDS
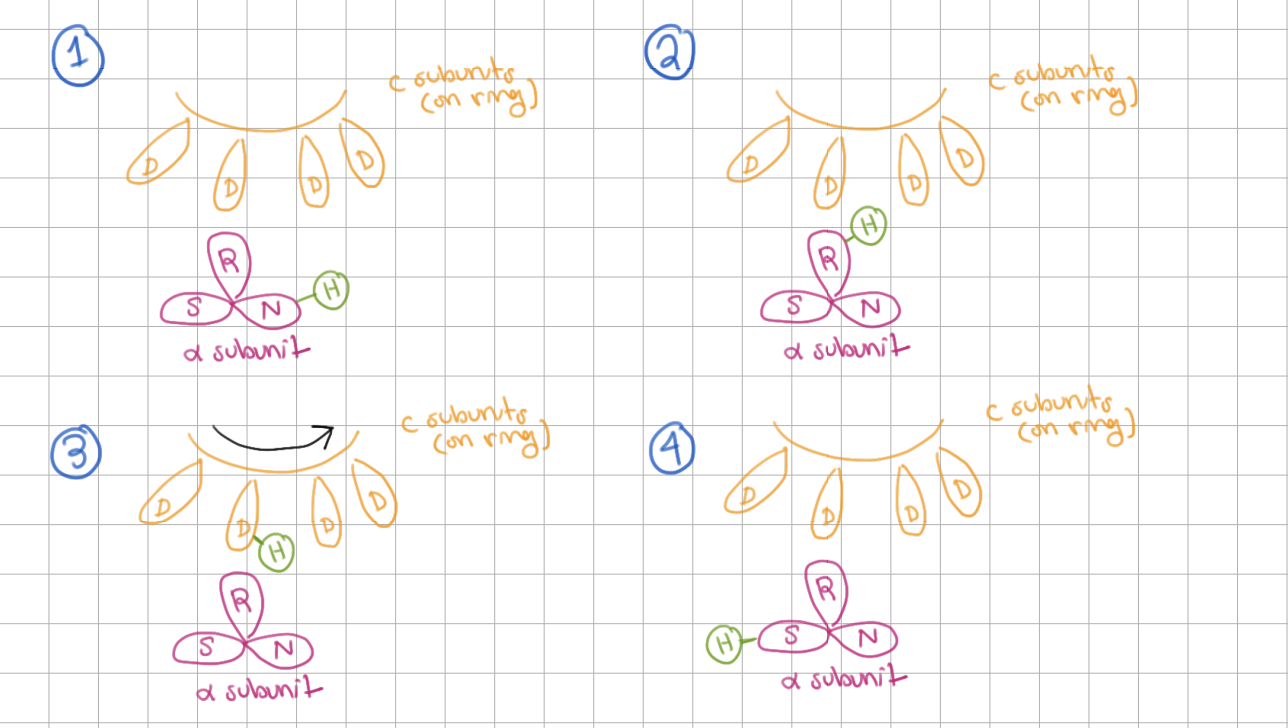
A and C Subunit Mechanism
H+ enters ___ at the __________
H+ hops onto ___
H+ goes to ___ on __________
H+ rides around rotor
H+ exits on ___ at the __________
A and C Subunit Mechanism
H+ enters N at the a inlet half channel
H+ hops onto R
H+ goes to D on c subunit
H+ rides around rotor
H+ exits on S at the a outlet half channel
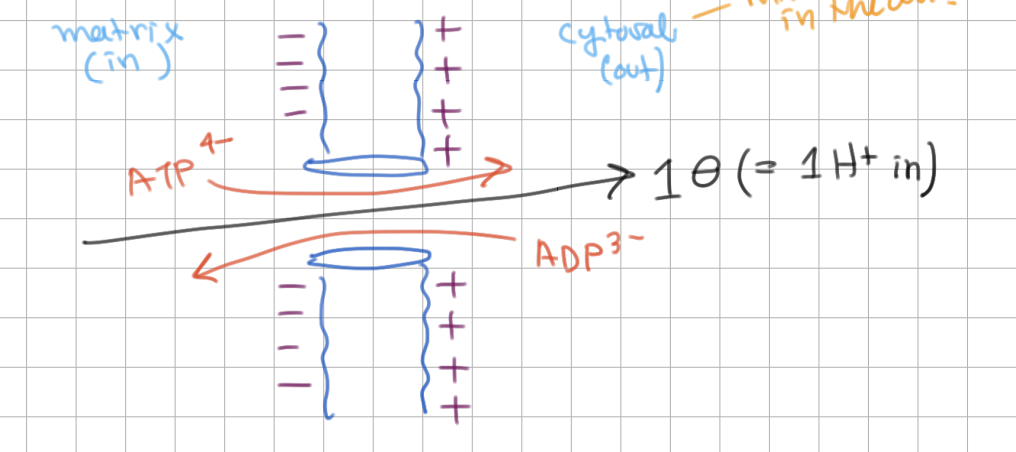
ATP-ADP Translocase in Mitochondria
ATP is only transported in/out
ADP is only transported in/out
ATP-ADP Translocase in Mitochondria
ATP is only transported out
ADP is only transported in
Cytosol (outside) is more positive/negative, so ATP movement out is spontaneous.
ATP movement is equivalent to 1 electron leaving/entering or 1 H+ leaving/entering.
-
Thus, the total cost of making and exporting ATP is ___ H+ (in).
Cytosol (outside) is more positive, so ATP movement out is spontaneous.
ATP movement is equivalent to 1 electron leaving or 1 H+ entering.
-
Thus, the total cost of making and exporting ATP is 4 H+ (in).
P/O =
# ATP / electron pair
ATPase with 10c subunits yields ____ ATP,
ATPase with 8c subunits yields ____ ATP
ATPase with 10c subunits yields 3.33 ATP,
ATPase with 8c subunits yields 2.7 ATP

Bacteria have no mitochondria, how does this affect ATP yield?
higher ATP yield (lose no H+ to export ATP)
