Chapter 12-Endomembrane System
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
37 Terms
Trafficking
Movement of lipids and proteins between organelles
Components of the endomembrane system
Endoplasmic Reticulum (protein synthesis)
Endosomes (Carry + sort material brought into the cell)
Lysosomes (digest ingested material and uneeeded cellular components)
Endoplasmic reticulum
The initial steps of addition of carbohydrates to glycoproteins
The folding of polypeptides
Recognition + removal of misfolded proteins
Assembly of multimeric proteins
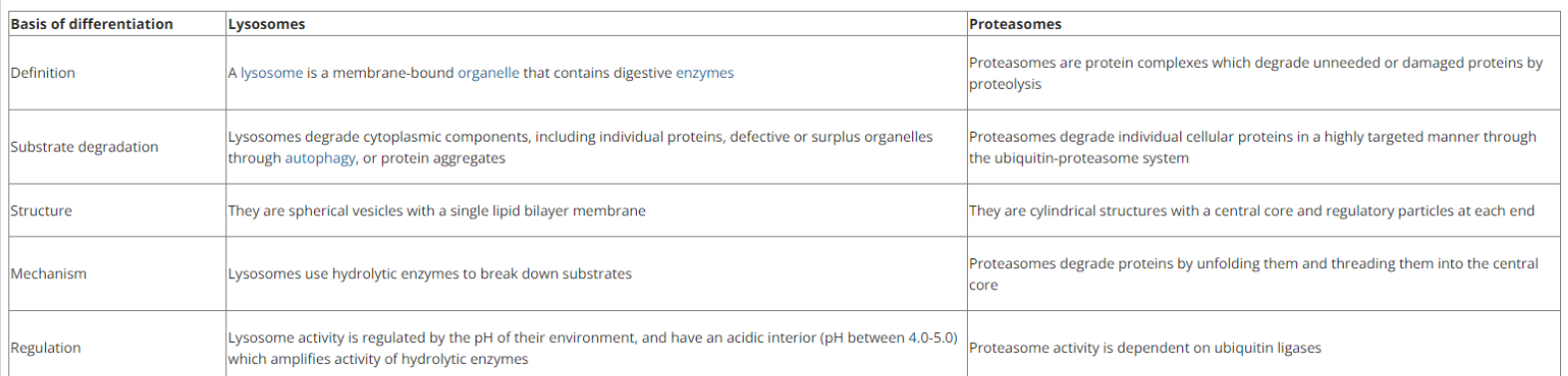
Proteasomes
Located in the cytosol
Degrades incorrectly folded, modified, or assembled proteins
A highly sophisticated protease complex
Lysosomes Vs. Proteosomes
Rough ER Vs. Smooth ER
Rough = Protein Synthesis
Smooth = Lipid Synthesis
Variation in Amounts of Rough and Smooth ER
Rough ER = Synthesis of secretory proteins
Smooth ER = Steroid hormone production
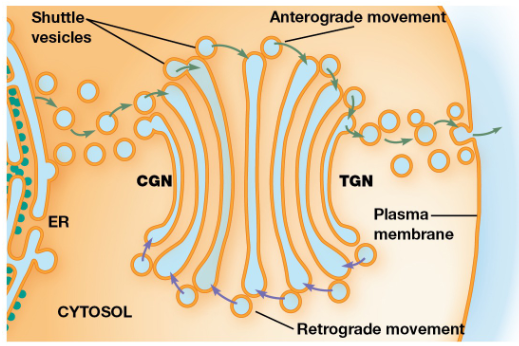
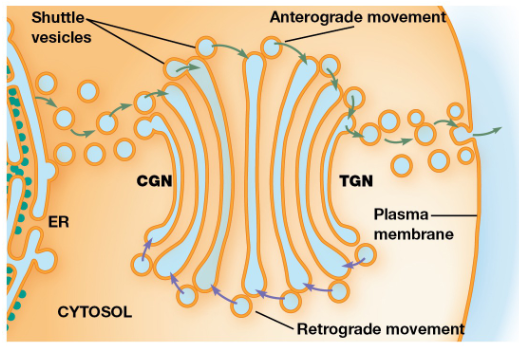
Golgi Apparatus
Glycoproteins & membrane lipids from the ER undergo further processing and are sorted + packaged for transport
Series of flattened membrane-bound cisternae (usually 3-8)
Cis, Medial, & Trans Golgi
cis-Golgi network = Oriented towards the ER
medial cisternae = Between the TGN and CGN
trans-Golgi network = Opposite side of the golgi
Stationary Cisternae Model
Each cisterna in the golgi stack is a stable structure. Transport of materials goes from cis-to-trans
Cisternal Maturation Model
The golgi cisternae are transient and they gradually change from CGN to TGN and enzymes are returned to earlier compartments
Anterograde Transport
Movement of material toward the plasma membrane
Retrograde Transport
The flow of vesicles from the golgi cisternae back to the ER
Protein processing in the ER & golgi apparatus
Glycosylation
The addition of carbohydrate side chains to proteins (forms glycoproteins that help serve to modulate the function of a protein. Can be the addition of nitrogen or oxygen)
Molecular Chaperones
Proteins that assist others to fold properly during or after synthesis, to refold after partial denaturation, and to translocate to the cellular locations at which they reside and function
Bi P
Binds the hydrophobic region of a polypeptide which prevents proteins aggregating together. It releases the polypeptide accompanied by ATP hydrolysis which allows
Protein disulfide isomerase
Disulfide bonds begin forming before synthesis of the
polypeptide is complete and bonds are formed/broken until the most stable arrangement can be found.
Unfolded Protein Response (UPR)
uses sensor molecules in the E R membrane to detect misfolded proteins. The sensors activate signaling pathways that shut down the production of protein except for those required for protein folding and degradation
ER Associated Degradation (ERAD)
recognizes misfolded or unassembled proteins and exports them to the cytosol to be degraded by proteasomes.
Protein Tags
Targets proteins to a transport vesicle that will take it to the correct location. A tag may be an amino acid sequence, a hydrophobic domain, oligosaccharide side chain, or some other feature
Pathway 1 Sorting Polypeptides
Cytosol, mitochondria, chloroplast, preoxisome, & nuclear interior
Free ribosome > Remains in cytosol or taken up by organelle
Pathway 2 Sorting Polypeptides
Endomembrane system or export
Polypeptide is transferred across or inserted into the ER membrane as synthesis occurs (cotranslational import)
Posttranslational Import
the uptake of completed polypeptides with special targeting signals into the organelle.
Cotranslational Import
The polypeptide is transferred across (or for integral
membrane proteins, inserted into) the ER membrane as
synthesis occurs
Membrane-enclosed organelles import proteins by one of 3 mechanisms
Transport through nuclear pores
Transport across membranes (proteins unfold)
Transport by vesicles
Nuclear Pore Complex
forms a gate through which selected macromolecules and larger complexes enter or exit the nucleus
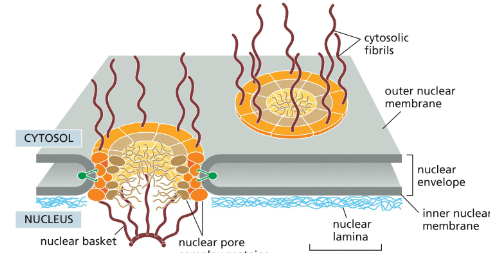
Prospective Nuclear Protein Transport
proteins contain a nuclear localization signal that is recognized by nuclear import receptors, which interact with the cytosolic fibrils
receptors with their cargo jostle their way through the gel-like meshwork until nuclear entry triggers cargo release
After cargo delivery, the receptors return to the cytosol
via nuclear pores for reuse
Ran-GAP
found in cytosol; converts Ran-GTP to Ran-GDP
Ran-GEF
found in nucleus; Ran-GDP > GTP
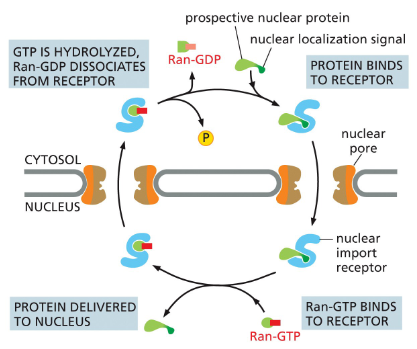
Steps of Nuclear Transport (Powered by GTP Hydrolysis)
A nuclear import receptor binds to a nuclear protein in the cytosol.
The receptor + protein complex enters the nucleus.
Inside the nucleus, Ran-GTP binds to the receptor.
This binding causes the receptor to release the nuclear protein.
The receptor, still carrying Ran-GTP, exits the nucleus back into the cytosol.
In the cytosol, Ran hydrolyzes its GTP → GDP.
Ran-GDP detaches from the import receptor.
The receptor is now free to pick up another nuclear protein.
Ran-GDP is returned to the nucleus by its own import receptor
Precursor Proteins
an inactive protein or peptide that can be turned into an active form by post-translational modification
Mitochondrial Protein Transport
A mitochondrial precursor protein has a signal sequence.
The signal sequence is recognized by a receptor on the outer mitochondrial membrane.
The receptor is connected to a protein translocator, which moves the signal sequence into the intermembrane space.
The complex of receptor + precursor protein + translocator moves sideways in the outer membrane.
The signal sequence is then recognized by a second translocator in the inner membrane.
The two translocators work together to transport the protein across both membranes, unfolding the protein as it passes through.
In the mitochondrial matrix, a signal peptidase cuts off the signal sequence.
Chaperone proteins (using ATP) help pull the protein across and assist with refolding.
Polyribosome
Groups of many ribosomes that bind to each mRNA molecule in the cytosol to synthesize a protein
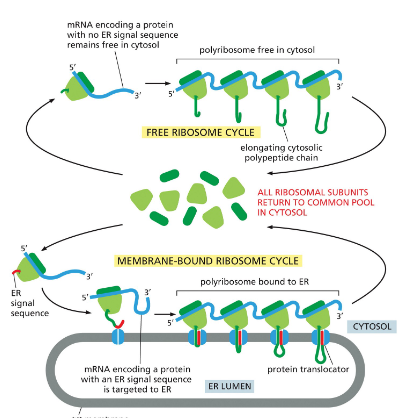
Directing a ribosome to the ER Membrane
The signal recognition particle (SRP) binds to both the exposed ER signal sequence and the ribosome.
This binding slows protein synthesis by the ribosome.
The SRP–ribosome complex binds to an SRP receptor in the ER membrane.
The SRP is released.
The ribosome is transferred from the SRP receptor to a protein translocator in the ER membrane.
Protein synthesis resumes, and the translocator threads the growing polypeptide chain across the ER membrane into the lumen.
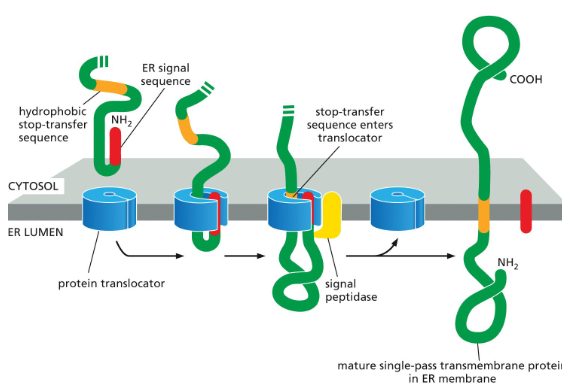
Single-pass Transmembrane Protein
The protein has an N-terminal ER signal sequence (red).
This signal sequence starts transfer into the ER through a protein translocator.
The protein also has a second hydrophobic sequence (orange).
This is the stop-transfer sequence.
When the stop-transfer sequence enters the translocator, it halts transfer through the channel.
The translocator then releases the growing polypeptide sideways into the lipid bilayer.
The N-terminal signal sequence is cleaved off.
The result is that the protein becomes anchored in the ER membrane, with one domain sticking into the ER lumen and the rest staying in the cytosol.
Protein synthesis continues on the cytosolic side until the protein is fully made.
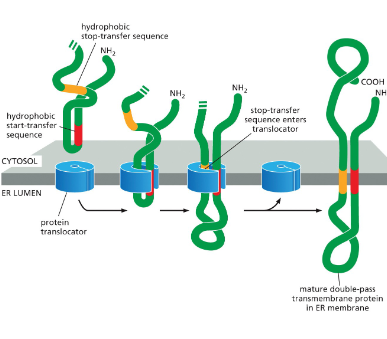
Double-Pass Transmembrane Protein
The protein has an internal signal sequence (red).
Acts as a start-transfer signal (lets the chain enter the ER).
Also acts as an anchor to hold part of the protein in the membrane.
This internal signal sequence is recognized by the SRP, which brings the ribosome to the ER membrane.
Translation continues until a stop-transfer sequence (orange) enters the protein translocator.
When the stop-transfer enters, the translocator releases both sequences (start + stop) sideways into the lipid bilayer.
Unlike the N-terminal signal in single-pass proteins, here neither the start-transfer nor stop-transfer is cleaved.
The result: the protein is anchored in the membrane at two hydrophobic stretches — creating a double-pass transmembrane protein.
For proteins that span the membrane more than twice, the polypeptide contains extra start/stop pairs, and the process repeats for each pair.