molecular bio
1/92
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
93 Terms
Molecular Biology
the study of gene structure and function at the molecular level
Gene
sequence of DNA that codes for a protein
Central Dogma
Gene expression occurs in two major steps - transcription and translation.
Gene Expression Steps
Genes (sequences of DNA), are inherited. DNA is replicated during cell division so that each somatic cell of an organism contains the same DNA (gametes, or reproductive cells undergo meiosis, and so they contain less DNA). DNA undergoes the first step, transcription, to make mRNA. Translation is the second step, and this is used to make protein.
How did we learn that DNA exists?
Pre-historic people recognized the existence of heredity
Domestication of animals (8000 - 1000 BC)
Cultivation of plants (5000 BC)
Desirable and undesirable varieties/traits of animals and plants
could be selected
Particulate Theory
Aristotle felt that parts, or particles were passed from each parent to an offspring
Gregor Mendel
Studied how traits were inherited in pea plants units of inheritance were passed from parents to offspring
Factors
were responsible for inheritance
some unit of inheritance was being passed from parent to offspring
Next major advancement: inherited factors are located on chromosomes
it was known that gametes (reproductive cells) are haploid - the number of chromosomes in these cells are cut in half
Thomas Hunt Morgan
credited with establishing that chromosomes are responsible for inheritance through the use of his fruit fly experiments
He mated white-eyed males and red-eyed females.
Red eyes are wild-type. In the first generation from this cross, all flies are red-eyed, which is not surprising because red eyes are dominant.
But in the F2 generation, white eyes appeared only in the males.
Linked Inheritance
Genes that are located on the same chromosome are "linked" and are usually inherited together
Crossing Over
chromosomes can exchange genetic information during prophase I of meiosis - mixing up the traits that can be inherited
Traits that are inherited more frequently together are closer together on the chromosome
Friedrich Miescher
discovered the existence of DNA
"Nuclein" precipitated in acid, and could dissolve again in alkaline conditions (not like protein) - so he knew that this was not protein, but had to be a different type of molecule
Genetic Material
Must provide information responsible for traits
Must provide information for replication of an organism
Must itself be replicated
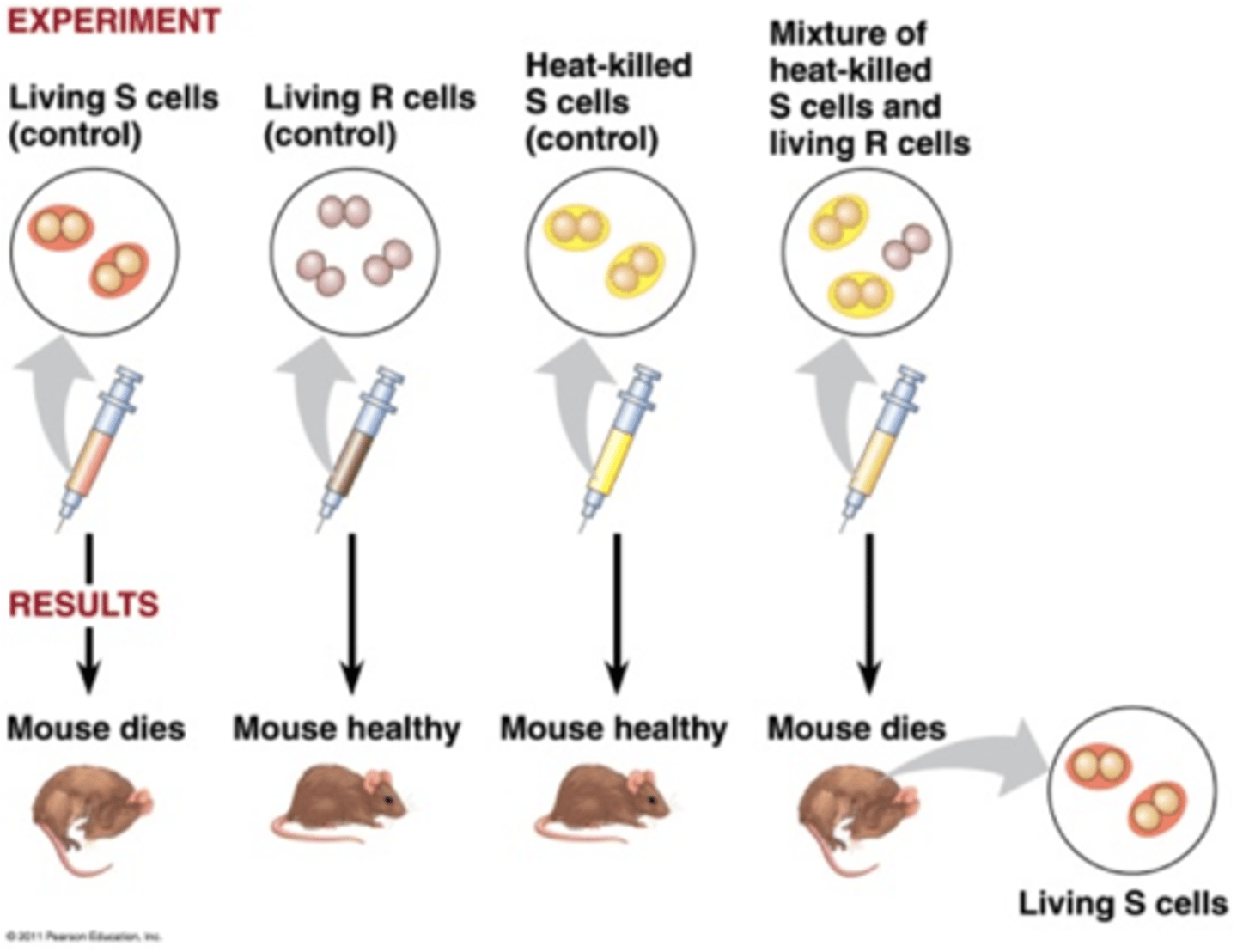
Griffith
He was studying the bacteria that causes pneumonia. One strain, S strain, had a smooth coat, and could therefore evade the immune system and was pathogenic in mice. The R strain had a rough coat, and does not cause disease.
Griffith Mouse Experiment
Mice treated with S: S causes disease, the mouse will die
Mice treated with R: R does not cause disease, mouse will live
Mice treated with heat-killed S: Heat will kill the bacteria - mouse will live
Mice treated with heat-killed S and live R: mouse dies.... Upon autopsy, both S and R living bacteria are found.... This could not be explained at the time it was done... it was explained years later
Avery
Explain Griffith
Isolated DNA from the S strain and added it directly to the R strain: the R strain was then "transformed" to become an S strain
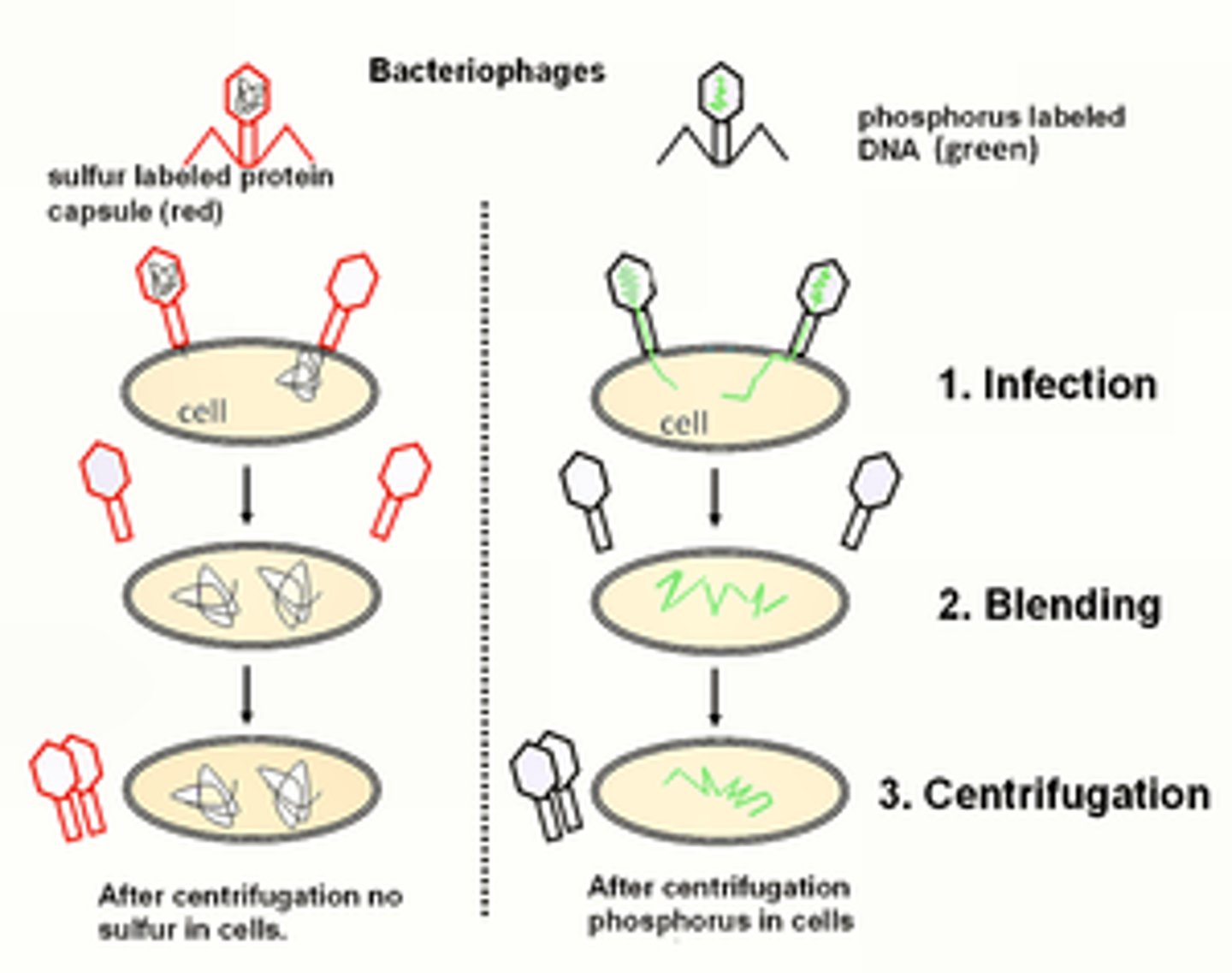
Hershey and Chase
On the left, the phage protein was labeled with S-35. The protein coat is shown labeled in blue as a result. The phage are allowed to infect, and then they used a blender to mix up and separate the phage from the infected bacteria, which could be centrifuged.
On the right, the DNA was labeled with P32. The labeled phages were allowed to infect, and the parent phages were again removed by blending and the infected bacteria were centrifuged.
They knew that the bacteria pelled were infected, and therefore would have offspring phages being made inside - what was responsible for the inherited traits of these phages? The could only detect P32 inside the infected bacteria, so DNA was responsible for inheritance of the phages.
Garrod
"inborn error of metabolism"
alkaptonuria
It was known that this was inherited as a recessive trait, and Garrod realized that there was one defective gene that caused this
So one defective gene resulted in one defective enzyme
Beadle and Tatum
one gene one enzyme
studying the growth of the mold Neurospora
Chargaff
first to realize that the nitrogenous bases of DNA were not found in equal number (A, G, C, T)
The #A's were equal to the #T's, and the #Cs were equal to the #Gs
He did not know about complementary base pairing, but he knew that the ratios existed
Franklin and Wilkins
X-ray diffraction studies of DNA - isolating DNA, hitting it with an X-ray to see the pattern of its structure
Watson and Crick
Could figure out that DNA had a helical structure.
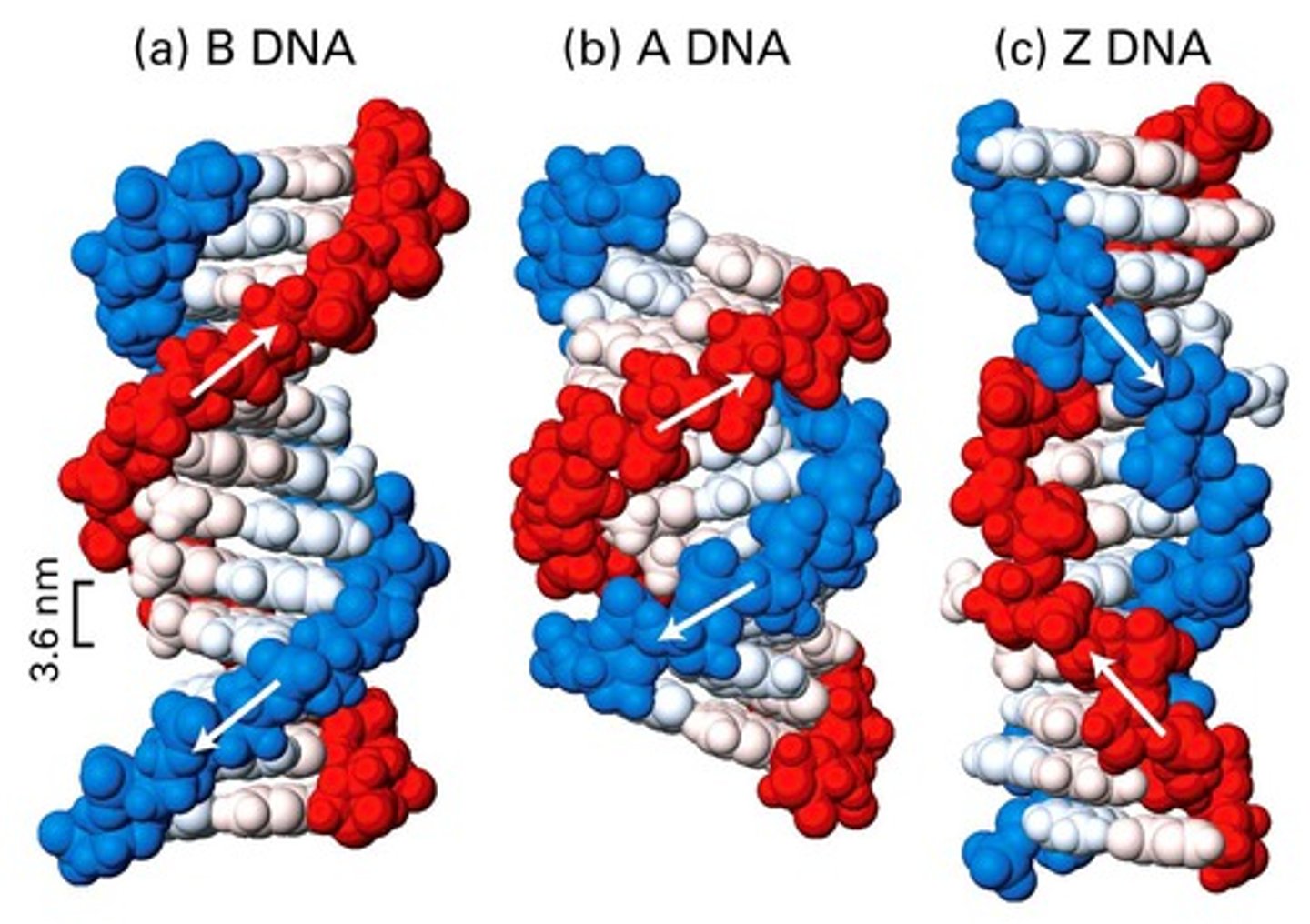
B Form of DNA
proposed by Watson and Crick
high humidity (92%)
Base pairs are horizontal
right-handed - the highest phosphate-sugar chain begins on the right and wraps around to the left
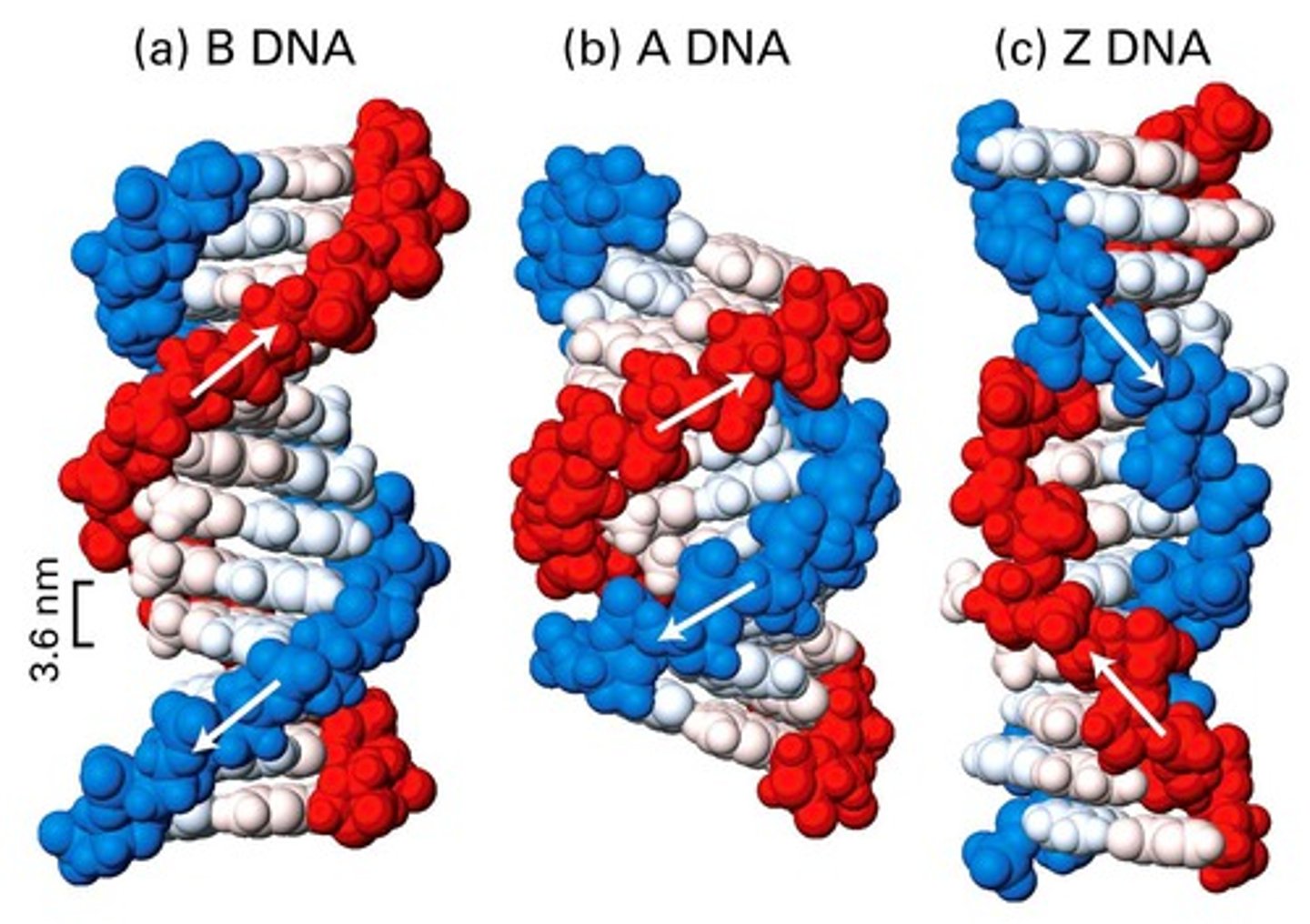
A-DNA
right-handed
found at 75% relative humidity
Plane of the base pairs tilts 20 degrees from horizontal
11 bp per turn instead of 10 bp per turn
each turn occurs in 31A instead of 34A
found in hybrids of DNA and RNA, dsRNA
Z-DNA
alternating purines and pyrimidines
least compact; function not known
left-handed
Linear DNA
DNA is usually found as a closed structure (lacking free ends) - even if it is linear
linear DNA is held looped out on a protein scaffold, but it is held in the scaffold - keeping it a closed structure
Circular DNA
found in bacterial and viral genomes and in eukaryotic cells in large loops
Supercoils
introduced into DNA when it is twisted around its own axis, can only happen in closed structures
Molecules that are linear or that lack supercoiling: relaxed
Separating Double Helix
harder to separate G's and C's
The easiest way to accomplish this is by using heat since heat can break hydrogen bonds. Denaturing DNA is also called "melting" DNA
Tm
the temperature at which half of the DNA strand is melted
This is greatly influenced by GC content
DNA Light Absorbance
How much DNA is melted can be determined by measuring absorbance at 260 nm. DNA absorbs light at 260 nm, and does this more efficiently when it is single-stranded. In this graph, you can see that as the temperature increases, the absorbance at 260 nm goes up
Other Factors that Influence Tm
Organic solvents:
High pH:
Lowering the salt concentration:
Organic SOlvents
functional groups can form H-bonds with the nitrogenous bases, preventing them from hydrogen bonding with each other
High PH
hydroxide ions (OH-) can form bonds with the hydrogens involved in hydrogen bonding between nitrogenous groups, pulling them apart
Lowering the Salt Concentration
this can remove ions that block the negative charges on the DNA backbone, which causes the two strands to then repel each other
Annealing or Renaturation
heat it at 95 degrees and then just turn off the heat block and leave it overnight - it will gradually cool and renature
Factors that influence renaturation efficiency
Temperature
DNA Concentration
Renaturation Time
Temperature
best 25°C below the Tm
high enough = rapid diffusion of DNA
low enough = doesn't promote denaturation
DNA Concentration
higher the concentration = more likely to come in contact
Renaturation Time
longer time allowed for annealing = more annealing will occur
CoT Curve
Describe the renaturation of DNA
reassociation depends on time and concentration
CoT1/2
time and concentration it takes for ½ the DNA to be renatured
Falling CoT Curve
represent re-associated DNAs.
At the top of the curve, 0% is renatured, when the bottom is reached, 100% is renatured.
Complexity of DNA
DNA influences the concentration/time (Cot) needed for reassociation
The complexity is the amount of unrepetitive sequence in the DNA molecule
Highly Repetitive
short sequences of DNA
satellite DNA: found at centromeres
telomere sequences
(10% of mouse genome)
Moderately Repetitive
genes for ribosomal RNA
(30% of mouse genome)
Single-Copy
not repetitive
protein-encoding regions
Hybridization
Putting together two strands of different origin
(PCR, Northern/Southern blotting)
Same considerations as reassociation:
PCR
You need to anneal primers to template DNA
the primers need to be rich in GC content (especially at the 3' end, as this is the end that needs to be extended with new nucleotides during PCR)
The concentration of primers in the reaction needs to be high enough as well
the Tm of each member of the pair of primers should be similar, and the annealing step of the PCR reaction should be done at 25 degrees lower than the Tm of the primer
Southern Blots
blots detect the presence of a DNA sequence by using a tagged ssDNA probe to bind to it
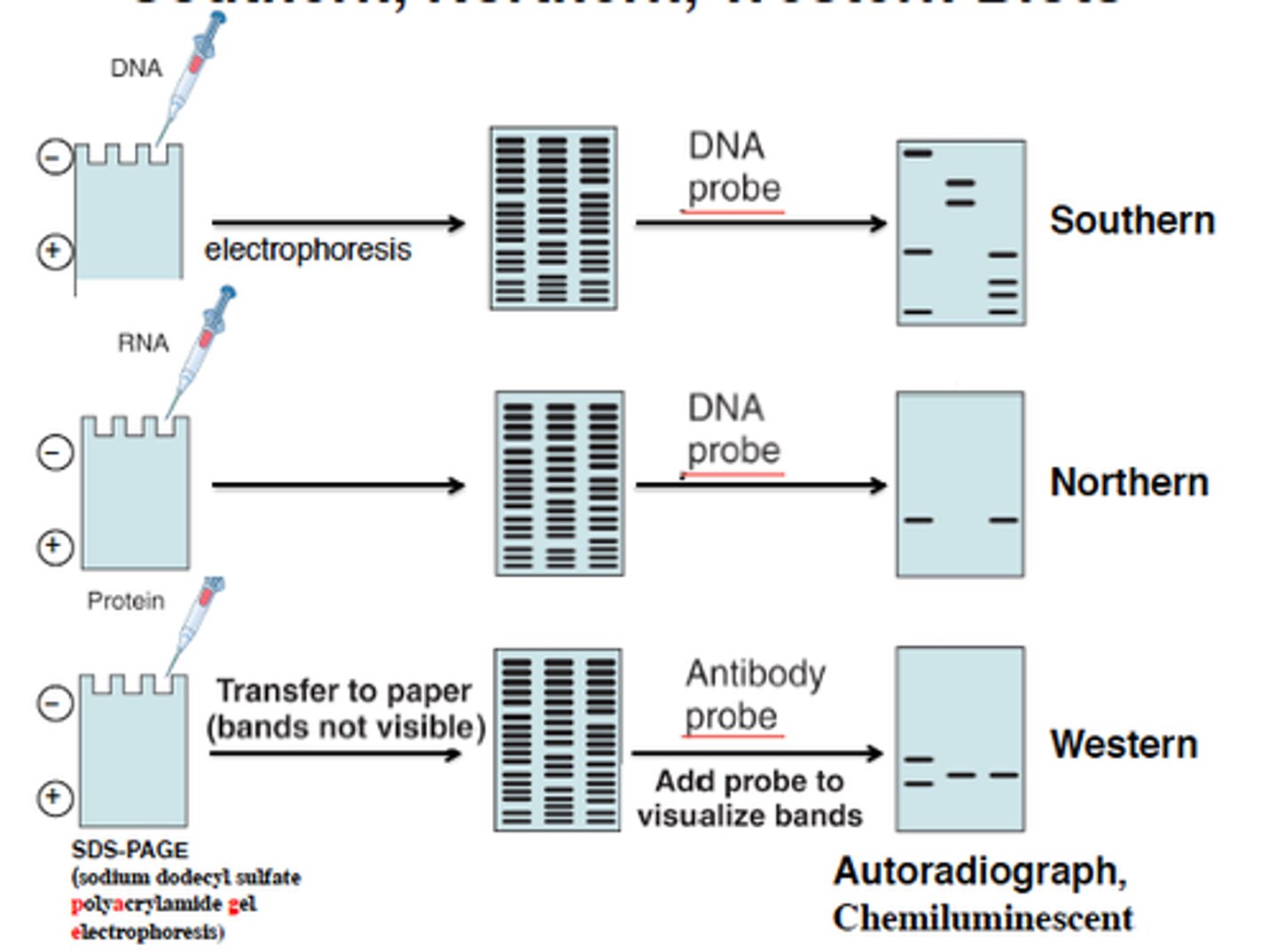
Northern Blots
detecting the presence of an RNA sequence by using a tagged ssDNA probe to bind to it
Blotting Considerationas
Concentration of probe needs to be high enough
Tm has to be considered when planning the experimental conditions: cannot incubate the blot too hot, or the probe will not bind to the sequence
Low Stringency
this allows maximum amount of probe to bind to sequences on the membrane
Base mismatches are more tolerated.
Typically it is done around 55 degrees - hot enough to allow movement, but low enough that a lot of probe can bind.
It is usually an overnight incubation (long time)
Washes
used to increase the stringency- making sure there is more specificity between the probe and the target
Unbound probe, and probe that is poorly matched to sequences on the membrane would be removed;
the washes gradually lower the salt and higher the heat - making it harder for non-specific interactions between the probe and target to be maintained
C-Value
DNA content per haploid cell
Measured in molecular weight, bp, and length
Histones
proteins with highly positively-charged regions
are 5 types of histones (H2A, H2B, H3, and H4 make up the nucleosome bead, histone H1 attaches next to the bead structure
histones appear as beads on a string.
DNA wraps around the nucleosome (146 bp of DNA fit), and then the H1 histone is added at the end
Additional Packing
This beaded string structure then undergoes additional levels of packaging.
It will fold to form 30 nm-chromatin fibers, which then fold more to form looped domains
Looped DOmains
fold to form condensed chromosomes, the folding makes the chromosomes visible during mitosis
DNA Replication
Replication is linked to the cell cycle:
1). Initiation commits the cell to further division
2). Division of the cell can not occur until DNA replication
is finished
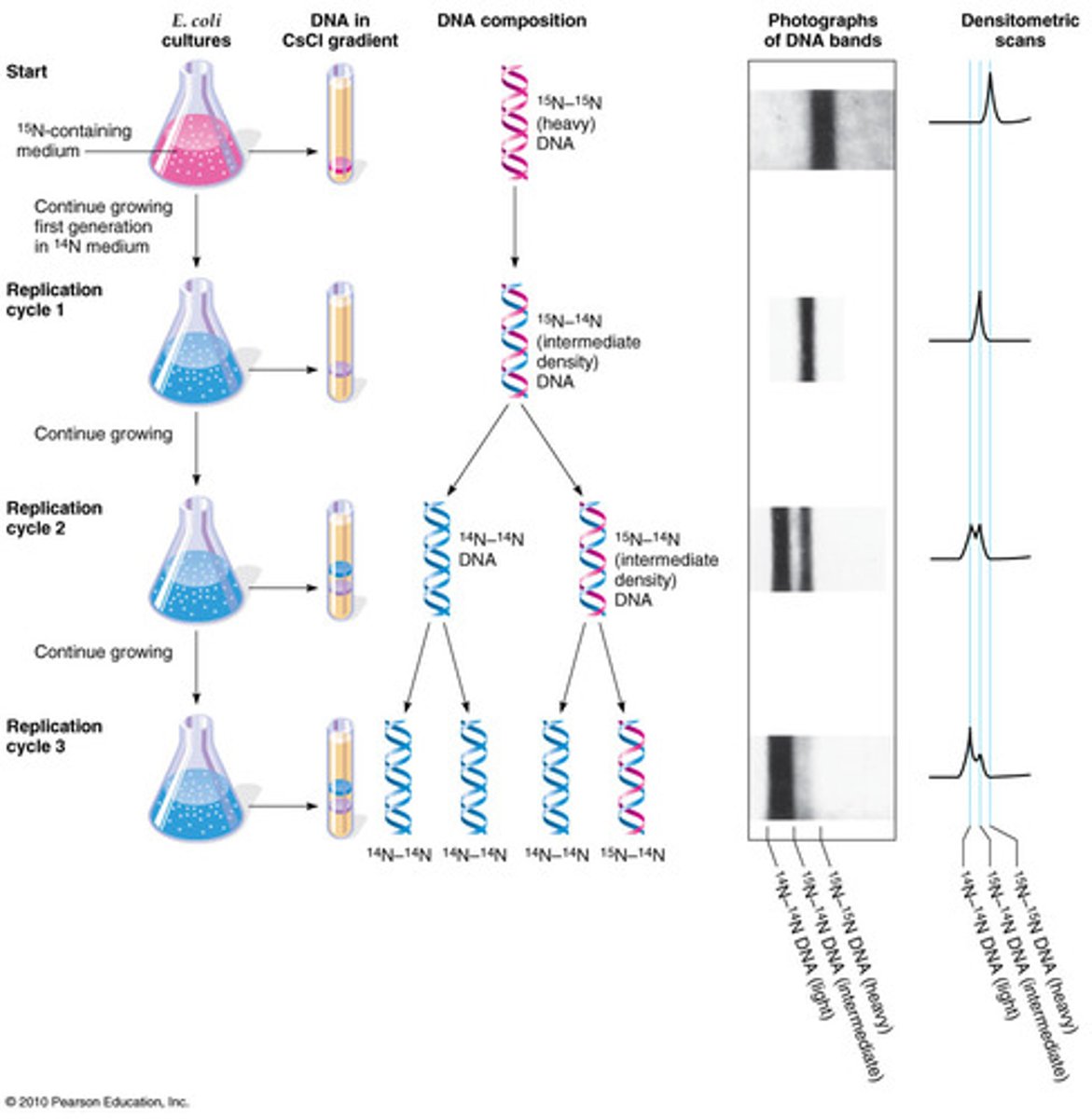
Meselson and Stahl
Proved that DNA replicates in a semiconservative fashion, confirming Watson and Crick's hypothesis. Cultured bacteria in a medium containing heavy nitrogen (15N) and then a medium containing light nitrogen (14N); after extracting the DNA, they demonstrated that the replicated DNA consisted of one heavy strand and one light strand
DNA Replication is Semidiscontinuous
DNA polymerase can only add nucleotides in a 5' to 3' direction... it needs a 3' end to attach the next nucleotide in the chain
However, when the fork opens on the right, the newly replicated piece at the top (shown in green) has a 5' end in the open region... DNA cannot be added to the 5' end... as the DNA unwinds further, the replication must be done backwards on this top strand, creating fragments (called Okazaki fragments) that are then linked together
DNA Ligase
an enzyme that eventually joins the sugar-phosphate backbones of the Okazaki fragments
DNA Replication Requires a Primer
DNA polymerase requires an existing 3' end to add nucleotides
Primer
short sequence of RNA (10- 12 bp) complementary to the DNA being replicated that is laid down first by a special type of RNA polymerase given the name "primase"
Replicon
Unit of DNA that is replicated from one origin of replication
Prokaryotic Genomes
single replicon
Plasmids
Extrachromosomal circular DNA
replicate autonomously
Eukaryotic Genomes
large number of replicons
each replicon is activated during S phase
Unidirectional
one replication fork
E. coli plasmid ColE1
Bidirectional
movement in two directions away from the origin
Some circular replicons, like the E. coli genome, are given the name "theta replication" because they look like the Greekletter theta, or an "eye" - this is bidirectional replication of a circular replicon
Eukaryotic Chromosomes
Replicate bidirectionally during S phase; replication continues along the replicon until it fuses with newly replicated DNA from the next origin; 15% of replicons are active at any given moment
Helicase
is the enzyme that separates DNA strands at the replication fork, and this enzyme requires ATP for its activity; multiple have been identified - in prokaryotes it is called DnaB
Single-Strand DNA Binding Proteins
; once the DNA is single-stranded, single-strand DNA binding proteins (SSBs) bind to it to keep it single stranded;
they bind cooperatively - meaning one binding helps the next one bind;
they may also be involved in recruiting DNA polymerase during DNA repair:
Topoisomerase
Introduces breaks into DNA allowing it to change ship
needed to change the shape of DNA
strand separation cannot occur without untwisting the DNA - DNA is not like a zipper with straight, parallel sides, but is a helix with strands
wound around each other
when the two strands of DNA separate, they must rotate around each other
•Closed circular DNA present special problems
-As DNA unwinds at one site more winding must occur at another site to reduce tension
Requirement for topoisomerases
Strands must unwind while they are separating
changes the topology of DNA
Negative supercoiling
(underwound,
less than 10.6 bp per turn)
Too few Turns
Positive supercoiling
(overwound, more than
10.6 bp per turn)
Too many turns
can prevent the polymerase from moving down the template if it is not corrected
Type 1 Topoisomerase
breaking one strand of double-stranded DNA
used to further relax negative supercoils, so these are not typically the ones needed during DNA replication
Type II Topoisomerase
DNA Gyrase in E. Coli
enzyme will bind one double-stranded part, and cut two strands, it will then pass another double-stranded section through it, relaxing the twisting
Cipro
works by blocking DNA gyrase activity in bacteria
DNA polymerase
adds triphosphate nucleotides to the replicating DNA by creating the phosphodiester bond
DNA Pol I
1). 5' to 3' polymerase activity - this is the main one we think about - it can add nucleotids to extend a 3' end - so it grows a new chain in the 5' to 3' direction
2). It has 3' to 5' exonuclease activity - this is also called "proofreading" activity - it can remove bases from the 3' end - this allows proofreading of mistakes made during replication - if a wrong base is added, it can be cut out and then replaced
3). It has 5' to 3' exonuclease activity - it can remove nucleotides from a free or nicked 5' end - this allows it to remove RNA primers - primer removal is the primary job of this particular enzyme; it is also involved in DNA repair
Klenow Fragment
: 5' to 3' polymerase activity and 3' to 5' proofreading activity
is used for primer extension DNA sequencing reactions... during sequencing, a primer is added and then you add Klenow fragment and nitrogenous bases that are labeled for detection (each nitrogenous base is labeled with a different color). Klenow will extend the primer and add labeled bases so that the sequence can be determined. You do not want the sequence to be degraded from the 5' end, which is why using just the Klenow fragment is a good choice for sequencing reactions. It can also proofread.
DNA Pol 1 Enzyme
Used for nick translation
5' ® 3' polymerase and 5' ® 3' exonuclease activity
both needed
The DNA pol I will use the 5' to 3' exonuclease activity to chop out bases from the 5' end
Then, DNA pol I will extend the 3' end at these empty regions by adding new nucleotides (some radioactively labeled) that are complementary to the opposite strand; DNA pol I cannot link the new fragment of DNA to the existing 5' end, so a nick remains. The nicks change location because of the activity of DNA pol I, which is why they called this procedure nick translation
DNase 1
Causes nicks in plasmid
DNA Pol 2
not needed for replication
Maybe a backup
DNa POl 3
3 subunits, 10 polypeptides
"replicase"
5' ® 3' polymerase activity
3' ® 5' exonuclease activity (proofreads
DNA polymerase d
replicates genomic DNA in eukaryotes
greatest processitivity
PCNA: clamps polymerase to template
Initiation
Requires a primer
Initiation In Prokaryotes
Occurs at the origin of replication
AT-rich
OriC: 9-mers = repeated regions
DnaA proteins are considered to be initiation factors for replication and will recognize the 9'mers and bind to the origin
DnaA
binding of DnaA recruits DnaB - DnaB serves as the helicase, and helps, along with HU protein, to separate the strands of DNA; HU protein has a structure similar to a histone, so it can bind to DNA and help to introduce negative supercoiling, making it easier for DNA to be separated