☠️🌐 Apoptosis + Endomembrane System
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
Mitochondria & Programmed Cell Death
Programmed cell death (apoptosis) is a normal process where cells die in a coordinated sequence
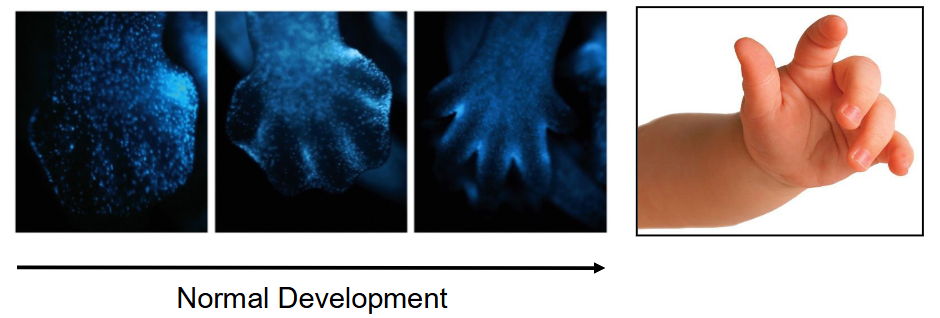
Part of organism growth/development
Example: Interdigital cell death causes soft tissue regression between embryonic digits in many vertebrates
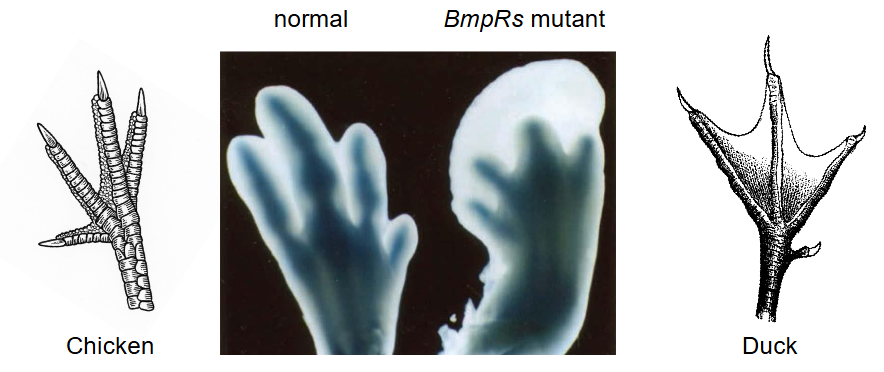
Bone Morphogenetic Protein (BMP) and Apoptosis
Bone morphogenetic protein (BMP) is a secreted protein that binds to Bmp receptors (BmpRs)
Expression of non-active BmpRs in chicken embryonic hind limbs:
Greatly reduced interdigital apoptosis
Results in webbed feet
Ducks don’t have mutation → Have webbed feet
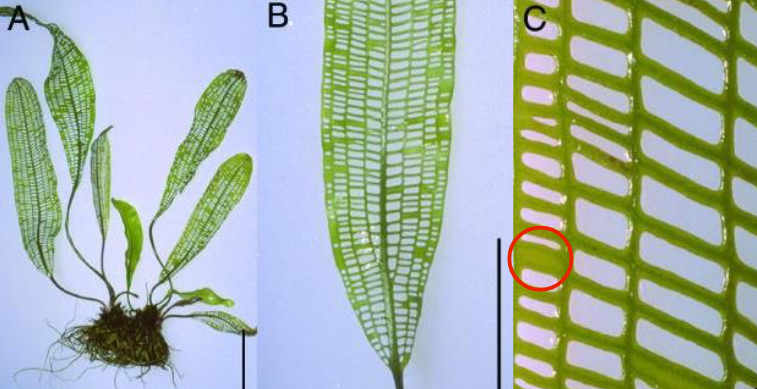
Apoptosis in Plant Growth
Apoptosis plays a role in plant growth
Madagascar Lace Plant:
A type of submerged aquatic plant
Has mature leaves that are fenestrated (contain holes)
Plant uses programmed cell death to generate holes in its leaves
Normal vs. Apoptotic Cells
Apoptosis is characterized by:
Shrinkage of cell
Blebbing (bulge/protrusion) of plasma membrane
Fragmentation of DNA and nucleus
Loss of attachment to other cells
Engulfment by phagocytosis
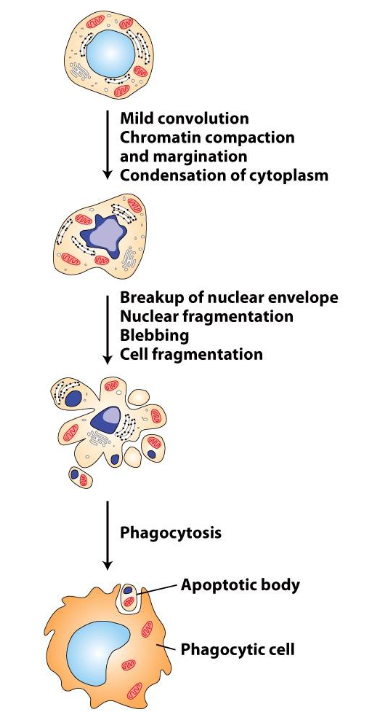
Steps:
Mild convolution, chromatin compaction, and margination
DNA begins to unwind out of chromatin and clumps against the nuclear envelope
Margination: Organelles are pushed to one side of cell
Condensation of cytoplasm
Cytoplasm shrinks and becomes more compact
Breakup of nuclear envelope
Nuclear envelope begins to break down
Nuclear fragmentation
Nuclear material fragments into smaller pieces
Blebbing
Plasma membrane bulges or protrudes
Cell fragmentation
Cell breaks apart into smaller apoptotic bodies
Phagocytosis
Apoptotic bodies are engulfed by phagocytic cells
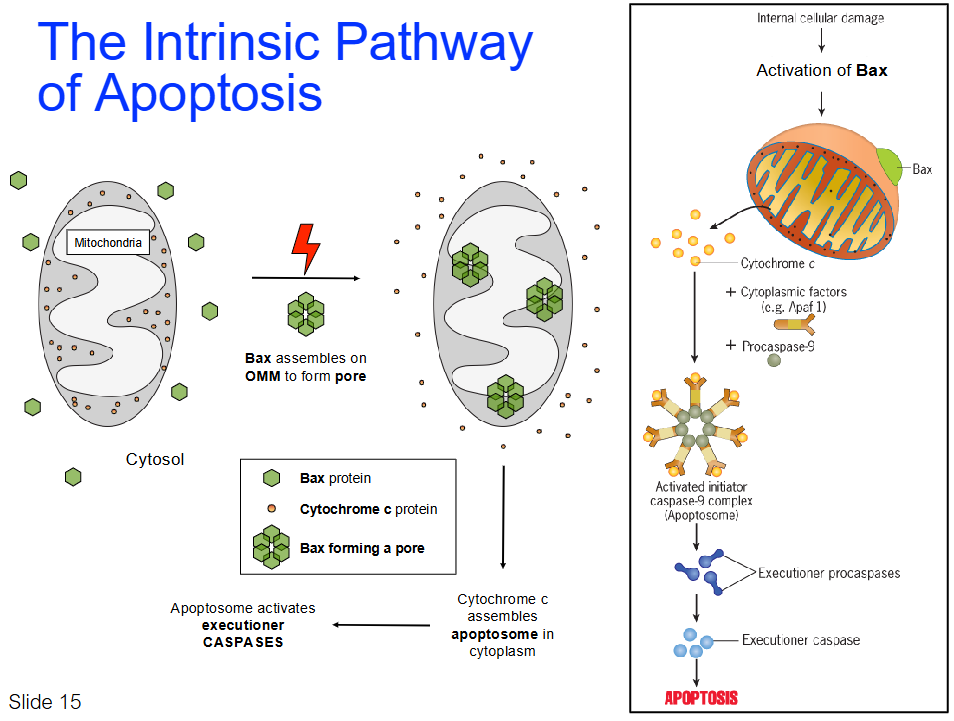
The Intrinsic (Initiated Internally) Pathway of Apoptosis
Initiated by intracellular stimuli (e.g., genetic damage, hypoxia, virus)
Killer proteins (e.g., Bax) cause changes in mitochondrial membrane potential (creates pores)
Bax proteins cause changes in mitochondrial membrane potential, leading to leakage of Cytochrome c
Bax assembles on outer mitochondrial membrane (OMM) to form a pore
Cytochrome c is released into cytosol
Apoptosome is formed by Cytochrome c and other proteins
Apoptosome activates executioner caspases, leading to apoptosis
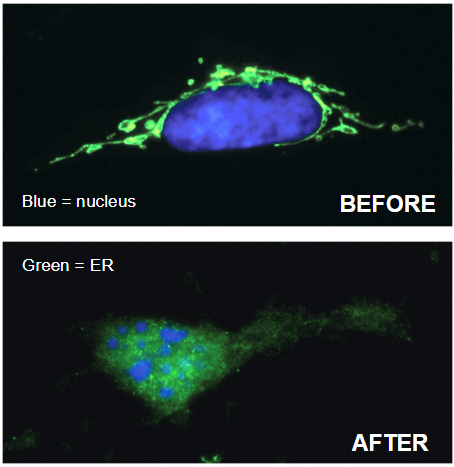
Release of Cytochrome c and Nuclear Fragmentation
Disrupts cell adhesion
Destroys lamins (nuclear filaments)
Breaks down cytoskeleton
Activates DNase (genome breakdown)
Before vs After:
Before: Intact nucleus and cell structure
After: Fragmentation and breakdown of cytoskeleton and nucleus during apoptosis
Caspases: Activated to carry out apoptosis
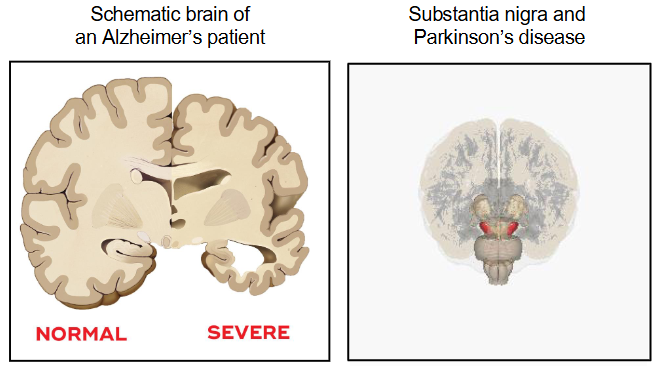
Apoptosis and Diseases
Various diseases are directly associated with apoptosis
In some cases, insufficient apoptosis leads to diseases like cancer (cells evade apoptosis, leading to uncontrolled cell growth)
In other cases, excessive apoptosis causes diseases like neurodegenerative disorders (e.g., Alzheimer's, Parkinson's) where too many cells die, leading to tissue damage
Main Functions of Intracellular Compartments
Cytosol: Protein synthesis, many metabolic pathways
Nucleus: Contains genome, DNA, RNA synthesis, ribosome assembly
Endoplasmic Reticulum (ER): Synthesis of lipids, synthesis of proteins
Golgi Apparatus: Protein modification, packaging of proteins and lipids
Lysosomes: Degradation of cellular material
Endosomes: Sorting, recycling
Mitochondria: ATP synthesis, apoptosis
Chloroplasts: Photosynthesis, ATP synthesis
Peroxisomes: Oxidation of toxic molecules
Early Electron Microscopy Observations of Cytoplasm
Membrane-Bound Organelles Identified by Early Electron Microscopy
Endoplasmic Reticulum (ER)
Endosomal Transport Vesicles
Golgi Complex
Lysosomes
Vacuoles
Early EM of cytoplasm revealed:
Membrane-bound organelles and vesicles
Extensive network of membranous canals
Stacks of cisternae (sac-like structures)
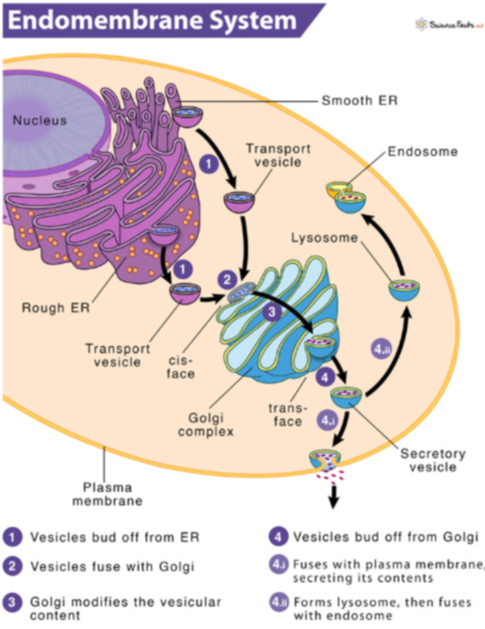
Polarized Structure of Secretory Cell
Secreted proteins (e.g. mucin, glycoprotein in mucus) are:
Synthesized in rough ER
Processed in ER
Further processed in Golgi body
Concentrated in vesicles
Delivered to plasma membrane for secretion and exocytosis
Goblet cells:
Produce mucigen granules (precursors of mucus)
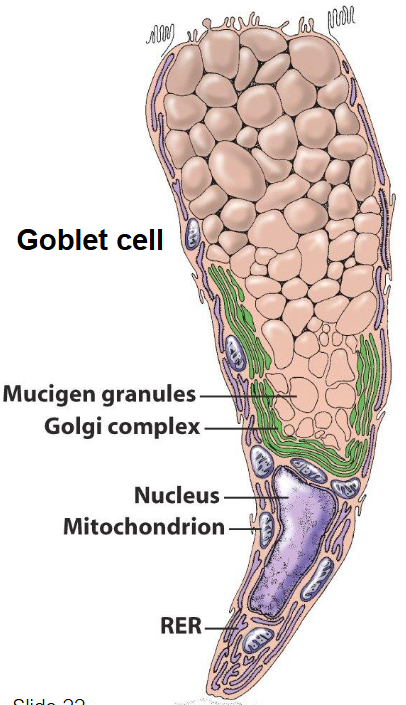
Overview of Biosynthetic / Secretory Endomembrane System
Nuclear envelope
ER
Golgi apparatus
Vesicles
Lysosomes
Plasma membrane (phospholipid bilayer + proteins, cholesterol, glycolipids, glycoproteins)
Facilitates:
Exo and endocytosis
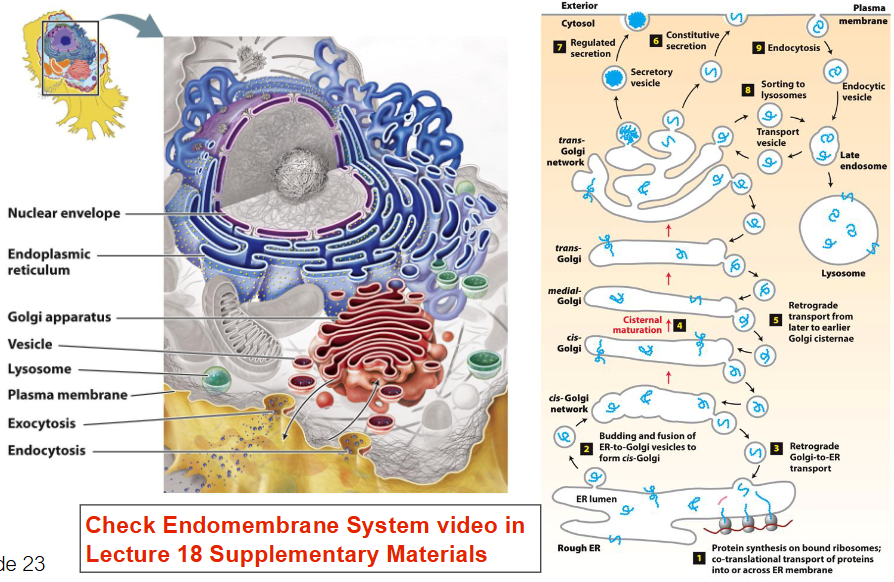
Protein Synthesis - ONLY KNOW RED
Rough ER & Translation
Ribosomes attached to rough ER synthesize proteins
mRNA from nucleus is translated by ribosomes
rRNA in ribosomes facilitates peptide bond formation
Proteins enter ER lumen during translation
In ER, proteins undergo folding and initial modifications
Transport Through Endomembrane System
Properly folded proteins are packaged into transport vesicles
Vesicles move from ER to Golgi apparatus
In Golgi, proteins are further modified, sorted, and packaged
Sorted proteins transported via vesicles to:
Plasma membrane (for secretion)
Lysosomes (for degradation enzymes)
Other organelles (for functional use)
Summary of Flow
Nucleus → mRNA
Ribosome on rough ER → translation
ER lumen → folding & modification
Vesicles → transport to Golgi
Golgi → processing & sorting
Vesicles → final destination (e.g. membrane, lysosome, secretion)
Technique: Using GFP to Track Cell Components
Green Fluorescent Protein (GFP) from Aequorea victoria (jellyfish)
GFP gene is fused with gene that codes for target protein
Fusion protein is expressed in cells
Allows visualization of protein's location, movement, and dynamics using fluorescence microscopy
Common tool in cell biology for studying protein localization, organelle tracking, and cell behavior
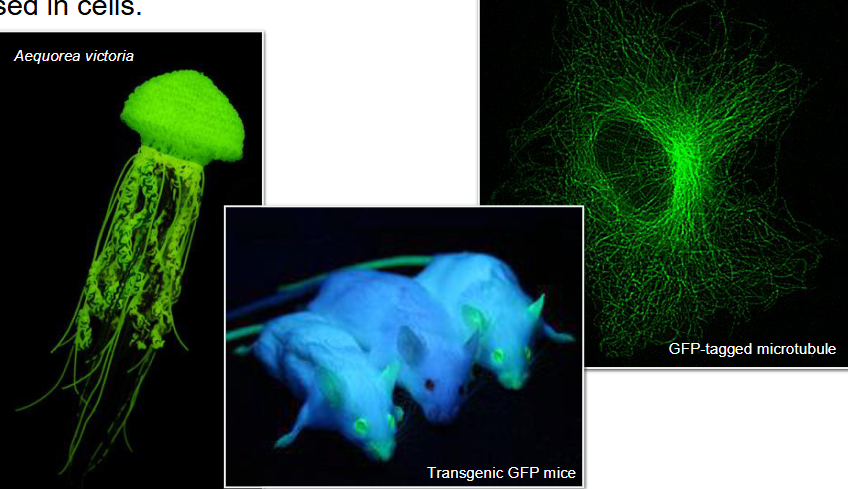
Using GFP to Track Cell Components
GFP fusion protein fluoresces, enabling visualization under a microscope
Observation of fusion protein reveals information about endogenous protein
Localization of protein in a cell or organism
Variants of GFP emit fluorescence at different wavelengths
Achieved through genetic modifications
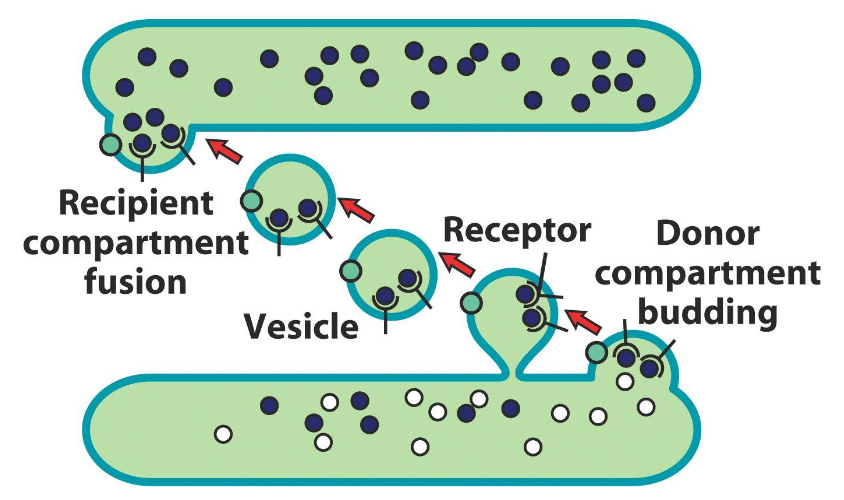
Transport of Material Between Compartments
Organelle → PM (and vice versa)
Organelle → Organelle
Uses transport vesicles (~50-75 nm)
Small, spherical, membrane-enclosed organelles
Bud off donor compartment and fuse with acceptor compartment
Targeted movement (directed)
Uses cytoskeleton and motor proteins
Sorting signals recognized by receptors
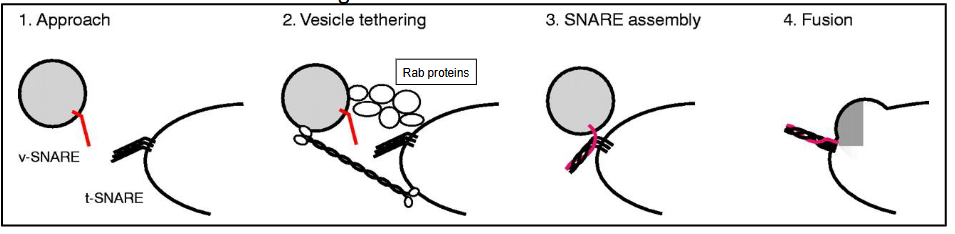
Key Elements of Vesicle Trafficking to a Compartment
Approach
Uses cytoskeleton and motor proteins
Can be anterograde (forward) or retrograde (backward)
Tethering
Uses Rab family proteins and other specialized proteins
Docking (SNARE assembly)
Vesicle has v-SNARE
Target membrane has t-SNARE
SNARE proteins interwind to form SNARE complex
SNARE complex pulls vesicle and target membrane closer (initiates lipid mixing)
Fusion of vesicle and target membrane
Fusion allows for cargo to release
SNARE complex is dissembled
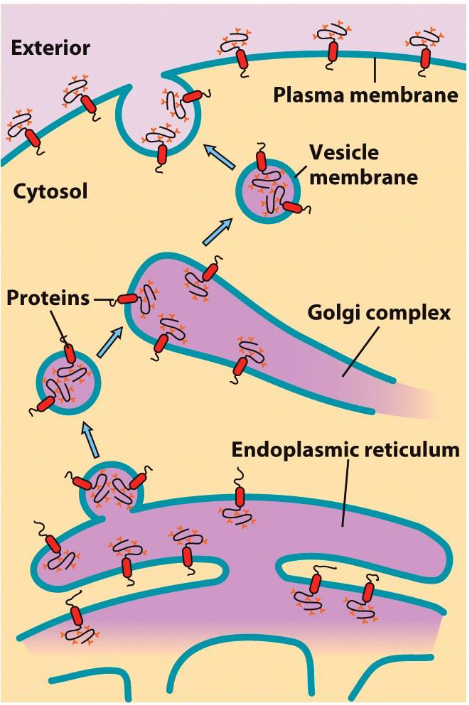
Protein Transport Through Endomembrane System
0 min: Protein starts in ER (where it’s synthesized)
40 min: Proteins are concentrated in Golgi apparatus for further processing
180 min: Proteins are transported to PM (to carry out functions)
Orientation of Transmembrane Proteins
Orientation of transmembrane protein is maintained through its travel
Cytoplasmic end of protein sticks out into cytosol
ER lumen end of protein faces into lumen of ER/Golgi/vesicle
*After exocytosis, ER lumen end of protein is on outside of plasma membrane since it merges and inverts
Proteins are tagged with signals to allow them to go to their designated locations
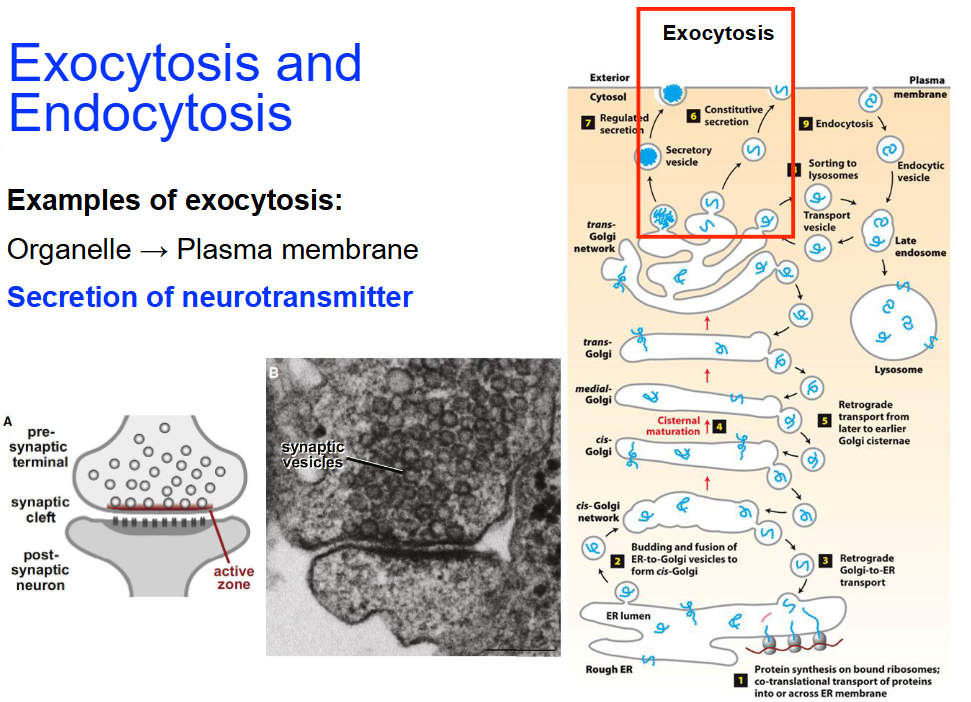
Exocytosis Examples
Organelle → Plasma membrane
Secretion of neurotransmitter
Regulated exocytosis (by Ca2+)
Process:
Action potential reaches the axon terminal, triggering calcium ion influx
Calcium ions facilitate the fusion of synaptic vesicles with the presynaptic membrane
Neurotransmitters are released into the synaptic cleft and bind to receptors on the postsynaptic membrane
This binding initiates a signal in the postsynaptic cell
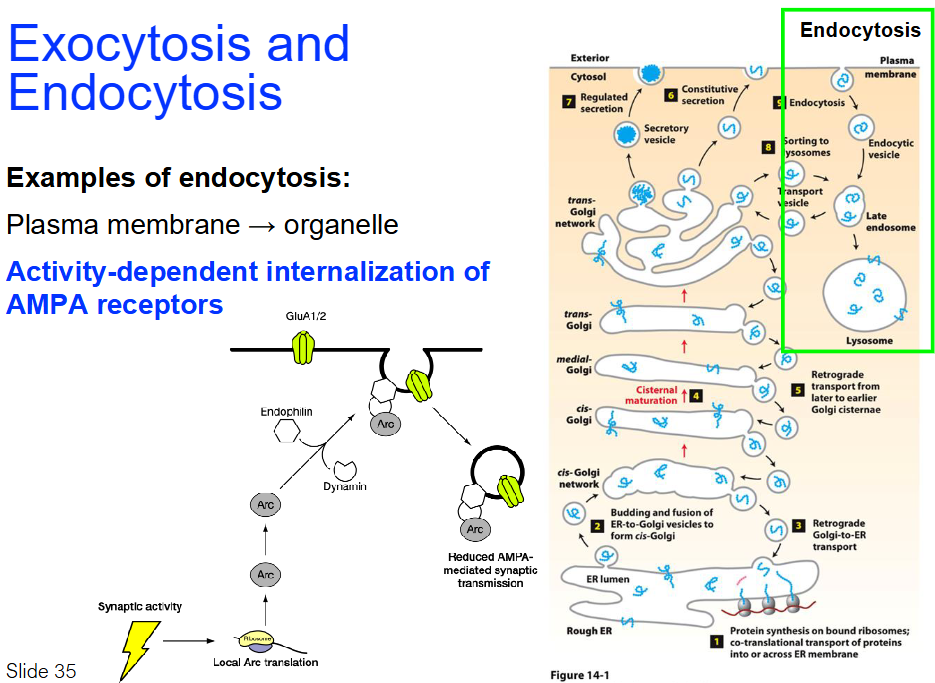
Endocytosis Examples - Highlighted is most important
Plasma membrane → Organelle
Reduces number of AMPA receptors on plasma membrane
→ Less Na+ enters neuron when signal arrives
→ Weaker synaptic response
Arc protein
→ Controls how many AMPA receptors are in membraneActivity-dependent internalization of AMPA receptors
GluA1/2: Subunits of AMPA receptors involved in synaptic transmission
Endophilin: Protein helps in vesicle formation during endocytosis
Arc: Protein involved in synaptic plasticity, translated locally at synapse
Dynamin: Helps vesicle fission during endocytosis
Reduced AMPA-mediated transmission: AMPA receptor internalization reduces synaptic transmission
Synaptic activity: Activates pathways that promote AMPA receptor internalization
Ribosome: Local translation of Arc near synapse during activity