bimm 120 midterm 2 study guide
1/80
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
81 Terms
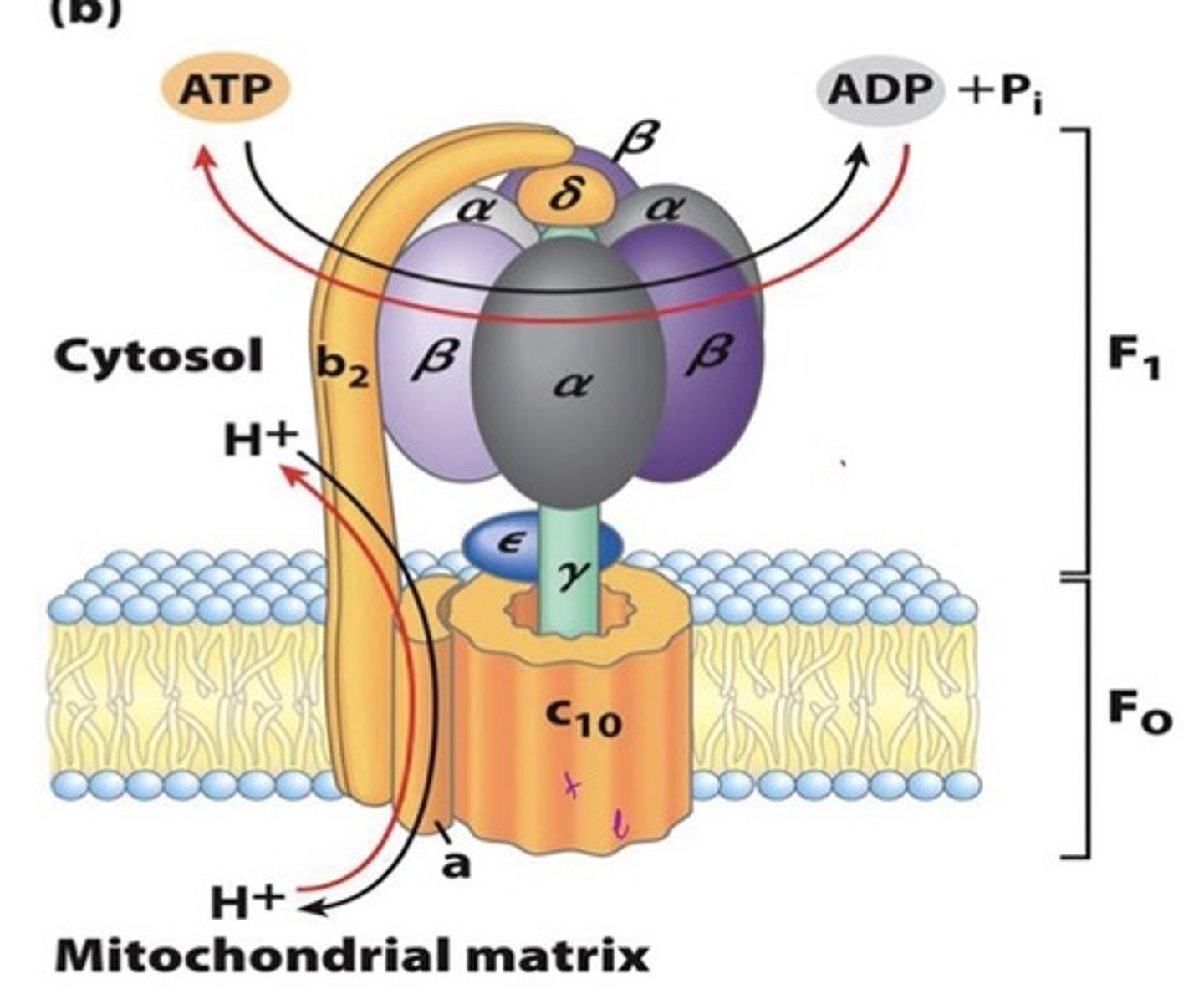
architecture of F-type ATPase
F1 = soluble, Greek
- alpha, beta, delta, gamma, and epsilon subunits
F0 = membrane-embedded, arabic
- subunits a, b, and c-ring
rotor: c-ring, gamma, and epsilon
stator: a, 2 b subunits, delta, 3 alpha, and 3 beta subunits
major components of bacterial flagellum
basal body: anchors the structure in the envelope/membrane and contains the (bidirectional rotary) motor
filament: extends many cell lengths from the cell surface and acts like a (helical) propeller
hook: universal joint that connects the basal body and filament
- build bottom up: basal body, hook, then filament last
environment and function of the flagellar rings
MS ring: membrane/secretory ring
- embedded in the inner membrane
C ring: cytoplasm ring
- in cytoplasm! not embedded in inner membrane
- associated with the motor (ring is the motor switch)
P ring: peptidoglycan-associated ring
- in periplasm!
- contains the stator = stabilized motor via anchoring
L ring: lipopolysaccharide-associated
- embedded in outer membrane
- connects to hook
FliC v. FljB switching in Salmonella
filament assembly in Salmonella will either be performed by FliC or FljB (only one expressed)
FljB & FliC are two distinct flagellin genes with domain that is recognized as H antigen
- FliC = H1 antigen, good for invading/colonizing tissue and tumbling
- FljB = H2 antigen, more flexible and mobile! better propeller for straight movement
Salmonella flagellin phase variation: switch between FliC and FljB to escape the host immune system by altering antigenicity
switching mediated by DNA inversion by Hin invertase
know the flagellar cap
HAP2 is the adapter at the tip of the filament
- guides incoming flagellin subunits to their terminal position
- causes the flagellum to stop growing
filament cap: FliD, keeps newly exported flagella from leaking out and chaperone for flagellin
what determines the power source for the bacterial flagellum?
the power source for bacterial flagellum depends on that availability of ions in the environment
H+ available: MotAB power source, H+-driven flagella
Na+ available: PomAB, power source is Na+ because lots in the environment
- PomAB used by marine Vibrio
- MotPS used by Bacillus (hyperalkalophilic)
what controls the lengths of the flagellar components?
FliK = ruler protein
- tells us when to stop building a flagellum
where is flagellin added/incorporated in flagellum?
under FliD!
flagellin units added to tip
Flip acts as chaperoning for flagellin by helping it refold and squeeze into the growing end
- also keeps newly exported flagellant from leaking out
key differences between bacterial and archaeal flagella
power source:
- archaeal flagella powered by ATP and have signal peptides
- bacterial flagella powered by pmf (H+ or Na+ in certain environments)
hollowness:
- archaeal flagella NOT hollow, monomers assemble at base
- bacteria flagella hollow = subunits flow through to assemble at tip
homologs: there are no homologous subunits or protein homologs between archaeal and bacterial flagella!
archaeal flagella homologous to type IV pilus
difference between flagella turning CCW vs. CW
flagella turning CCW = coherent swimming!
flagella turning CW = tumbling
how does the arrangement of flagella determine what happens when switching between CCW and CW rotation?
flagella bundle behind bacterium for CCW rotation = move forward
flagella disperse for CW rotation = tumble/go backward (rigid flagella)
what channel/motor powers flagellar rotation?
MotAB = H+ driven in E.coli and Salmonella
- ion selector
- C-terminal domain of FliG interacts with MotAB to produce torque
what are the three promoters involved in flagellar expression?
class 1: flhDC
- flhD and flhC = transcription factors that are controlled by global regulators, required to express subsequent promoters
class 2: HBB
- hook-basal body
- includes TFs FliA (turns on class 3 filament genes), FlgM (turns off class 3 genes until hook is ready)
class 3: filament but also some regulatory proteins
what are the different types of motility for prokaryotes?
swimming: flagella rotate together CCW to swim forward
swarming: with 1000 flagelli, they go faster because there's so many of them
pilus-mediated twitching: pilus extend from bacteria, bind to a surface, retract = move (grappling hook)
centipede/inchworm: cytoplasmic contractile filament proteins = actin in cytoplasm spread out then contract = movement
what are the different types of motility that are referred to as gliding?
flavobacterial gliding: adhesins bind to substratum/surface and help cell move slowly to LEFT via rotation of adhesins
myxococcal A-motility: polysaccharide secretion generates jet propulsion movement and leaves slime trail
why is oligomerization/polymerization useful with regards to genome economics?
if you need a lot of things to make various structures, but only have a small genome:
- express a lot of one thing (monomer) that can aggregate easily and form various structures quickly
feedback loop examples in microbio
changes in adhesion after attachment:
- attachment leads to virulence activity
- virulence leads to immobility
- immobility leads to biofilm formation
- biofilm formation = attachment & more virulence
positive feedback loop results in robust transitions to virulent states
why is chemotaxis so important?
attractants attract, repellants repel
- hydrophilic substances often attract (yum, sugars and amino acids!)
- hydrophobic substances often repel (oil only good in small amounts)
chemotaxis = ability to swim toward or away from chemical
common/general properties of chemoattractants and repellants
chemoattractant = hydrophilic substances often attract bacteria
chemorepellant = hydrophobic substances often repel bacteria
two-component systems
receptor + response regulator = MCP + CheY in chemotaxis
MCP = receptor for chemoattractant or chemorepellant
CheY = response regulator
- if phosphorylated, response regulator is active = CW turn (random movement)
- if inactive = CCW turn
difference between activation and adaptation
activation (phosphorylation/phosphate hydrolysis): fast! attractant/repellant bind = phosphorylation of CheA = sensor kinase active, etc.
adaptation (methylation/demethylation): slow, allows bacteria to detect differences in concentration over time & expands measurable range of chemoattractant/repellant concentrations
what does adaptation do for bacteria?
allow it to sense gradients (rather than absolute concentration responses) and expands the detectable range
- detects differences in concentration over time (memory)
- plateauing attractant concentration = stop running and stick around
- decreasing repellant concentration = keep running away
CheB versus CheR
CheR = methylase, activity level never changes, constant, constitutive
CheB = only active when phosphorylated by CheA-P, demethylase
interplay between chemotaxis and collective motion at diff cell densities
above a certain density, swirls develop in cell swimming problems
- below threshold of cell density: neighbor motility helps smooth chemotactic motion
- above threshold: neighbor motility becomes self-reinforcing and interferes with chemotaxis
ie. fish schooling! there is a good point
how is replication initiated in E.coli, oriC?
1. DnaA binds to a series of of DnaA boxes = localized melt/opening of AT-rich DNA
2. DnaC and DnaB bind next to DnaA to form a larger complex
3. Helicase (DnaB) unwinds the DNA, creating a large bubble of single stranded DNA
- DnaC falls off once replication is initiated
4. ssDNA is coated by SSB (single stranded DNA binding proteins) to keep the sticky strands apart
5. DNA polymerase complex loads
how is DNA compacted to fit inside E.coli?
Mg2+ counterion: neutralize neg charge of nucleic acid backbone = compact DNA
histones: cancel out neg charge of DNA backbone to neutralize it and compact it
supercoiling: gyrase generates supercoils by breaking 2 bonds, grabbing the nicked DNA to prevent uncoil, and introduce new supercoil by passing one strand through the other (ATP-dependent)
what is the nucleoid? how does it differ from nucleus?
nucleoid = highly supercoiled circular chromosomes in bacteria
- contains all DNA (chromosomes, plasmids, etc)
- not a nucleus because there is no nuclear membrane
- nucleoid is blob of supercoiled chromosome in center of bacterial cell
- 1 or 2 chromosomes + plasmids tightly folded, can be many COPIES of the chromosomes
how does the hydrophobicity of a molecule constrain the location of its receptors?
surface v. intracellular:
- intracellular receptors for molecules that can cross the membrane (AHLs, hydrophobic molecules)
- surface receptors for molecules that cannot cross the membrane (hydrophilic molecules, NA)
what is NA and what are AHLs?
NA = noradrenaline
- small hydrophilic molecule that cannot cross the membrane
- released when human is stressed
AHLs = acyl-homoserine lactones
- lipid, can cross membrane
- induces proinflammatory expression patterns in mammalian cells
- immunogenic in plants
what are AHLs a common target of?
eukaryotic eavesdropping! other molecules, like pathogens, can detect and respond to chemical signals produced by the AHLs
- pathogens do this and become more virulent
what are the functions and compositions of bacterial organelles covered in class?
acidocalcisomes: universal, lots of Ca2+, H+, polyphospate
- energy homeostasis, osmoregulation
anammoxosomes: oxidize ammonium with nitrate to generate water and N2
- like bacterial mitochondria for plantomycetes ONLY
carboxysomes: fix carbon dioxide, - consist of rubisco & carbonic anhydrase
- CO2 trapped due to protein shell
(bacterial) chromatophores: arise from PM invagination, contains all the enzymes involved in photosynthesis, pigment-dense compartments in photosynthetic bacteria
magnetosomes: align cell poles to magnetic field, size always the same about
what minerals are magnetosomes made of?
magnetite (Fe3O4)
- iron and oxygen
- aerobic
greigite (Fe3S4)
- iron and sulfur
- anaerobic
what are the different types of nanowires?
pilin-based nanowires/electrically conductive pili = e-pili
- conductive in absence of native metal cofactors or added metals, long range e- transport
curli fibers = poor conductivity naturally, boost with aromatic AA insertion
- biofilm and sensing
protein wires from cable bacteria: cable bacteria in aquatic sediments form long chains of cells that shuttle excess e- from sulfide oxidation to oxygen-rich areas
- lose conduction when exposed to air
which bacteria and archaea tend to have/use nanowires?
bacteria: Geobacter sulfurreducens, Schewanella, Synechococcus, Pelomaulum
archaea: methanospirillum hungatei
which directions do electrons flow in nanowires? what process does this mimic?
electrons flow from inner membrane to periplasm to outer membrane
- electrons are transferred to external electron acceptors
mimics biological respiration, but for anaerobic microbes
what common features of nanowires distinguish them from common pili by making the nanowires conductive?
nanowires have a high content of aromatic amino acids = conductive, e- delocalization in aromatic rings
- close packing of aromatic residues = continuous electron-conducting pathways
how can we make use of nanowire-producing microbes or their nanowires?
used for bioremediation (cleaning up ground water), converting animal waste into electricity, harvesting pili for e- conduction, mutant pili used for biotechnology
what are the non-motility processes/functions flagella are involve in?
attachment: flagella adhere to surfaces, initial stage in biofilm development
- biofilm for survival and proliferation, protection from chemical and biological agents
- adhesion to surfaces helps with resistance to antibiotics by enhancing PM stability
mechanosensing: flagella act as touch/pressure sensors , detects proximity to a surface, aids with successful how colonization (virulence)
how does natural selection work?
what does it mean when a trait survives and is widespread?
growth v. search strategy in E.coli:
- flagella expensive to run, why run them when nutrients are scarce?
- search strategy: benefits of sampling more space to find nutrients outweigh metabolic costs on a species level
- individual cells can and do starve to death in search of carbon
- the luck cells that find carbon fix this behavior in species
- growth strategy: risks of sampling more space to find even more nutrients outweigh the benefits of staying around on a species level
- growth can only speed up so much !
what are the common parts required for signaling pathways to work?
receptors, second messengers, effectors/response regulators, transcription factors
what are bacterial capsules?
a slime layer beyond the bacterial cell wall that is protective & sticky
- bind to surfaces, can form aggregates, they can bind to medical implants (typically leads to infections...)
- avoids immune system because polysaccharide layer not recognized by antibodies, prevent antibodies from binding to other bacteria
spirochetes and their internal flagella
flagella of spirochetes are in the periplasm, so they swarm in viscous fluid
- snake-like bacteria, travel fast over viscous surface
carbon storage granules
store carbon for energy via granules
- glycogen
- PHB: polyhydroxybutyrate
gas vesicles
hollow tubes made of proteins that are impermeable to H2O, permeable to gases, and have rigid protein shell
- organelle of floatation
- float at right oxygen level
- for photosynthesis
nucleoid
A non-membrane-enclosed region in a prokaryotic cell where its DNA (chromosomes and plasmids) is located
cellulosomes
protein machine on surface of cell, external to cytoplasm/periplasm
- contains scaffolding, which has tons of degradative enzymes bound to it
carboxysomes
have protein shells to compartmentalize enzymes, found in ALL cyanobacteria, used for CO2 fixation & chemitrophic generation of energy
cohesin
domain in genome that binds to another domain found at the C-terminus of degradative enzymes
prokaryotic cytoskeletal proteins & their eukaryotic homologs
PhuZ: positions replicating bacteriophage DNA in the center of the cell
TubZ: highly divergent tubular relative that plays a role in plasmid segregation
ParM: bacterial actin, polymerizes to push plasmids toward opposite poles of cell
SopA: polymerizes at same rate for either chromosomal and plasmid segregation (actin)
FtsZ = highly conserved cytosolic GTPase that forms Z-ring for cell division in bacteria and archaea
key differences between gram-positive and gram-negative bacteria
gram-negative bacteria: inner membrane, periplasm, peptidoglycan layer, outer membrane
gram-positive bacteria: cytoplasm, plasma membrane, peptidoglycan layer (outside of cell)
differences between diff levels/classes of oxygen utilization
obligate aerobe: migrate to air-water interface, seek oxygen
obligate anaerobe: migrate away from air-water interface, flee oxygen
facultative anaerobe: prefer oxygen but can make do without it
microaerophile: seek specific oxygen concentration
aerotolerant: no strong preference for or against oxygen
rhodopsins (review)
rhodopsins activate phototactic systems through methylated chemotaxis proteins
- use light energy for ion transport (causes pKa changes)
what does the change in free energy mean for reversibility/spotaneity of a process? what does this mean for active transport?
if free energy (delta G) is < 0 = release of energy, irreversible, spontaneous
if free energy (delta G) is > 0 = non-spontaneous process, uses energy
- active transport = G>0
how do the different levels of oxygen utilization pertain to transporters, particularly secretion systems?
secretion systems need ATP, and if there is low oxygen, there is low ATP production
- oxygen detected via respiration (FAD) and proton motive force
how are the following sensors accomplished: aerotaxis, magentotaxis, thermotaxis?
thermotaxis: 4 MCP thermosensors
- Tsr detects warmth
- Tar detects cold and warmth
- Trg is warmth sensor
- Tap is cold sensor
aerotaxis: oxygen concentration detected via respiration (FAD oxidized detection) & pmf
magentotaxis: orient to Earth's magnetic field, bacteria move sideways, up & down, in response to geomagnetism
- polar flagella needed in bacteria
- ion channels open and close in eukaryotes
why do MCPs have multiple methylation sites?
MCP have multiple methylation sites for adaptation to broader range of attractant/repellant concentration:
CheR : regulated
CheB: busy, active when phosphorylated
how and why can Tar act as both a warm and cold sensor?
native Tar has to be processed to be thermosensitive
- when Gln hydrolyzed to Glu for all 4 MCP sites = run toward heat
when Glu are methylated = run toward cold
what do bacterial proteasomes do?
mark proteins for degradation
what are bacterial chaperonins?
finish protein folding for partially folded/misfolded proteins
chaperonins similarities and differences with proteases
similar: heptameric barrel-like assemblies, powered by ATP hydrolysis
difference: functions
- chaperonins: fold misfiled proteins
- proteases: marks proteins for degradation
what signal do bacterial chaperonins recognize, how does it differ from eukaryotic proteasomes?
bacterial chaperonins recognize proteins that are misfolded or that have exposed hydrophobic surfaces
eukaryotic proteasomes recognize ubiquitin tags
what is the role of ATP hydrolysis in proteasomes and chaperonins?
proteasomes: ATP hydrolysis powers spooling of substrates into proteasome interior
chaperonins: folding powered by ATP hydrolysis
how does liquid-liquid phase separation play a role in forming membraneless compartments in cells?
enzymes form hyperstructures, they do not just float in cytoplasm of prokaryotes
- enzymes demix and form dense, liquid-like droplets
what is the role of adhesins on virulence, especially in Heliobacter pylori and Streptococcus progenes?
Heliobacter pylori: susceptible to stomach cancer if one has fucose in stomach, which is detected by fimbrium (adhesin) & bacteria stick around = ulcer or cancer
Streptococcus progenes: strep throat, flesh eating
- has 2 surface adhesins that bind to cell surface proteins and keratinocytes = colonization of throat/skin
inner-membrane components of Type I secretion system
ABC = ATP-binding cassette translocase
- in inner membrane of type I secretion system
= ATPase that couples transport process to ATP hydrolysis
- MFP in periplasm
inner membrane components of Type II secretion systems
Sec/Tat
Sec = ubiquitous
- signal peptidases, hydrolyze signal peptide off protein being transported once it crosses cytoplasmic membrane
- SecY/Sec61alpha: transporter
- SecDF system: interconverts ATP/GTP v. pmf for power
Tat = not ubiquitous, but widespread
- uses pmf only
- TatC = energizer using pmf
- TatA forms channel through inner membrane through which the protein travels
- protein is cell wall lysin, which hydrolyzes cell wall of peptidoglycan for set-destruct or phage takeover
inner membrane components of Type III secretion systems
Fla/Path
- YscD, YscQ
- powered by ATP hydrolysis
- related to flagellin exporter
inner membrane components of Type IV secretion system
Conj/Vir
- Vir proteins = inner membrane
- powered by ATP hydrolysis
T-plasmids in the context of Agrobacterium tumefaciens infection
potential pathogenicity with plants: Agrobacterium tumefaciens transfers cancerous plasmid with transfer-DNA into plants
power sources for inner membrane parts of type I - IV secretion systems
type I: powered by ATP
type II:
- Sec powered by GTP or ATP+pmf
- Tat powered by pmf
type III: Fla/Path powered by ATP+pmf
type IV: Conj/Vir powered by ATP
what does MFP do?
periplasmic component of type I secretion system:
MFP = membrane fusion protein
- function: interlinks inner and outer membrane transport pathways
outer membrane parts of type I - IV secretion systems
type I: OMF/OMP = outer membrane factors/proteins
type II: MTB = main terminal branch
- or secretin
type III: secretin or flagellar LP rings
type IV: Vir
what's the difference between two partner systems and two-component systems?
two-partner secretion system: 1 substrate protein + 1 transport protein (neighboring genes, only in bacteria)
- large proteins with adhesive activities that are linked to bacterial virulence
- both secreted to periplasm via Sec, passenger domain further exported to outer membrane
versus receptor and response regulator
AT-1 and OmpIP as secretion systems
AT-1: autotransporter-1 system
- single protein with N-terminal Sec-type signal peptide, a central passenger domain and a C-terminal beta-barrel domain
- secreted by Sec, beta-barrel domain insertes into outer membrane and passenger domain can be released or attach to cell surface
OmpIP: outer membrane protein insertional porin
- gram neg bacterial OMPs assembled from periplasm into the outer membrane
- secreted by Sec, inserted into outer membrane
- Omp85 involved in lipid biosynthesis
- if deleted, inner membrane accumulates LPS, while outer membrane losses LPS = cell weak and dies
motility-related homologs of secretion systems
T3SS-flagella
- flagella v. needle
- needle injects directly into eukaryotic host's cytoplasm to inject toxins
- share: ATPase, M/S ring
T2SS-pili/archaella
- MTB related to type IV pili and archaeal flagella
- 14 protein constituents
- ATPase & its anchor/chaperone, integral inner membrane/periplasmic proteins, four prepilins, outer membrane secretin
why is Sec good for inferring phylogenies?
phylogeny matches history: archaea give rise to eukaryotes, bacteria are before archaea
- great diversity in bacteria
how are fimbriae built? how does that process differ from how flagella are built?
fimbrial usher protein system = encodes a fimbrium-specific periplasmic chaperone protein & one for outer membrane usher
- pilus subunits secreted by Sec to periplasm, bound to chaperone in periplasm, preventing self-assembly inside the cell
- usher forms pore & helps release subunits from chaperone, exporting them to assemble on membrane surface
function of co-transported chaperones, like fimbrial usher protein
FUP is responsible for biogenesis of numerous fimbriae/pili in gram-negative bacteria
intracellular v. extracellular pathogens
bacteria are outside host cells = extracellular
- attach via adhesins
- Helicobacter pylori
- E.coli
toxins enter host cells = intracellular
- enter with invasin help
- Listeria (hijacks actin rockets), Herpes
what role do adhesins and invasins play in infection and their differences?
adhesins: recognize and attach to surfaces, help bacteria stick to epithelial cells for H. pylori
invasin: allows invasion into host cell, essential for intracellular pathogen
what are other virulence factors?
proteases: break down proteins
toxins: lyse cells, stop critical cellular processes, induce aberrant signaling behavior