L15/16: Transcriptional and post-transcriptional mRNA regulation (nuclear)
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
43 Terms
What are the 3 main processes needed to produce a mature mRNA + what are 2 additional levels of control
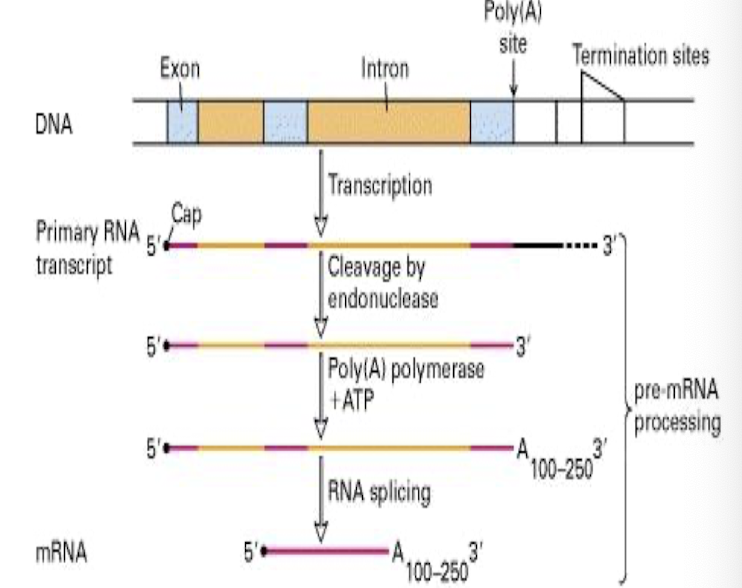
mRNA capping
Polyadenylation
Splicing - removal of introns
(Additional) Alternative splicing
RNA editing
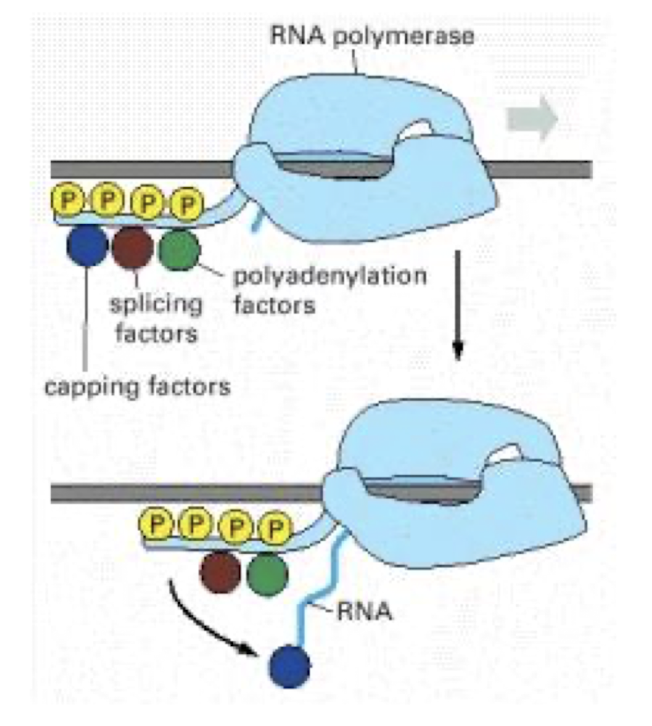
What is the importance of the RNAP II CTD for pre-mRNA coupling of transcription + mRNA processing events?
Carries pre-mRNA-processing proteins on its tail e.g. capping + splicing factors, which are transferred to the new RNA at the appropriate time
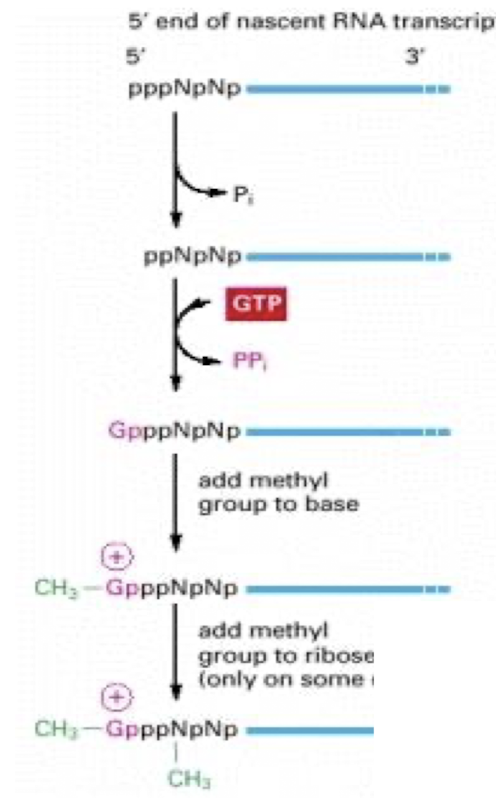
How is the 5’ Cap formed?
3 enzymes bind to phosphorylated Poly RNA tail & modify 5’ end of nascent RNA transcript as soon as it emerges from the polymerase
1. Phosphatase– removes P from 5’ end of nascent RNA
2. Quanyl Transferase adds a GMP in a reverse linkage (5’ to 5’ instead of 5’ to 3’)
3. RNA Methyl Transferase (RNMT) – adds CH3 group to the guanosine at position 7
What are the functions of the mRNA cap?
1) protects 5' end of mRNA from exonuclease degradation
2) binds CAP-Binding-Complex (CBC) facilitates nuclear export
3) Enhances translational efficiency
4) necessary for subsequent splicing
What is the significance of 3’ pre-mRNA polyadenylation?
It involves the addition of a poly(A) tail to the 3' end of pre-mRNA, by the addition of up to 200 adenosines
Occurs at poly A site flanked by 2 conserved sequence elements:
AAUAAA in 3’ UTR
GU-rich signal sequence
*Most eukaryotic mRNA have poly A tail EXCEPT histone mRNAs
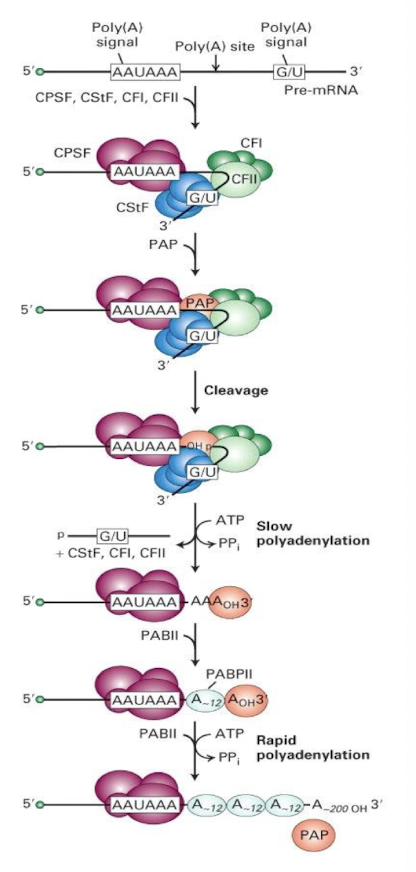
How can polyadenylation be summarised?
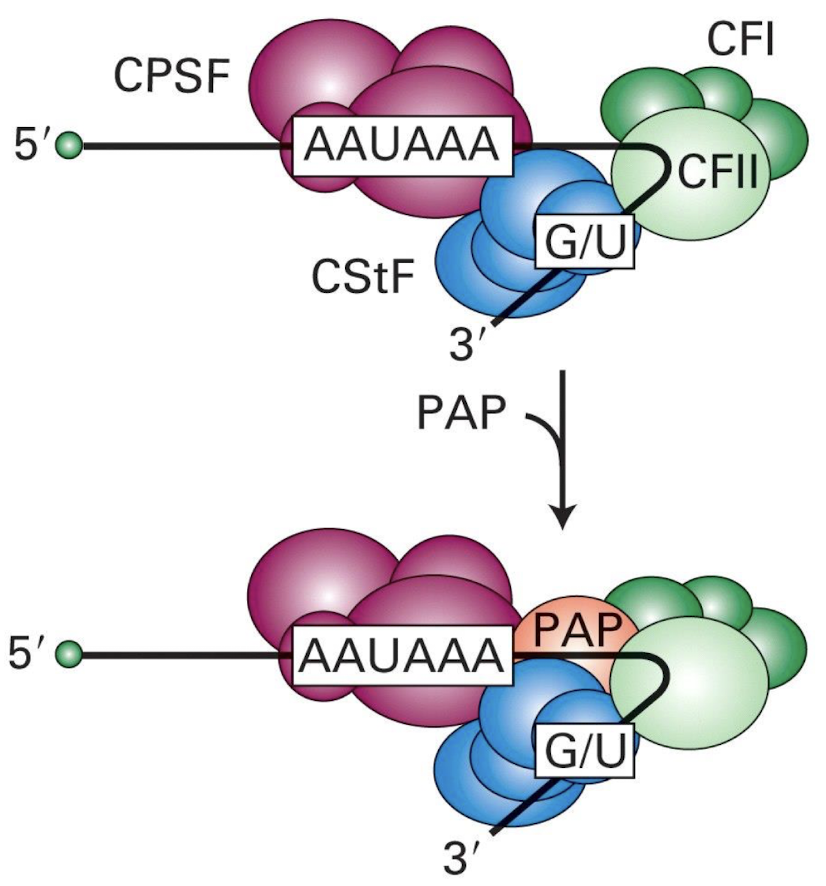
Multi-protein complex formation on mRNA e.g. by CPSF, CStf, CFI and CFII
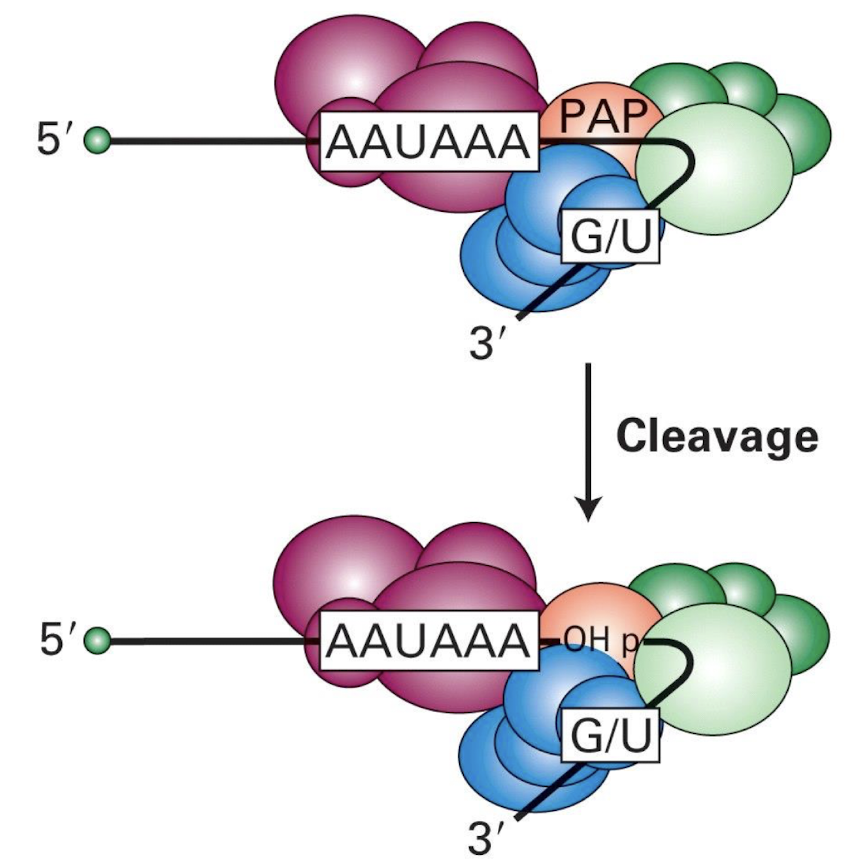
Binding of PAP prior to cleavage at poly A site → exposes OH group
Slow and rapid polyadenylation
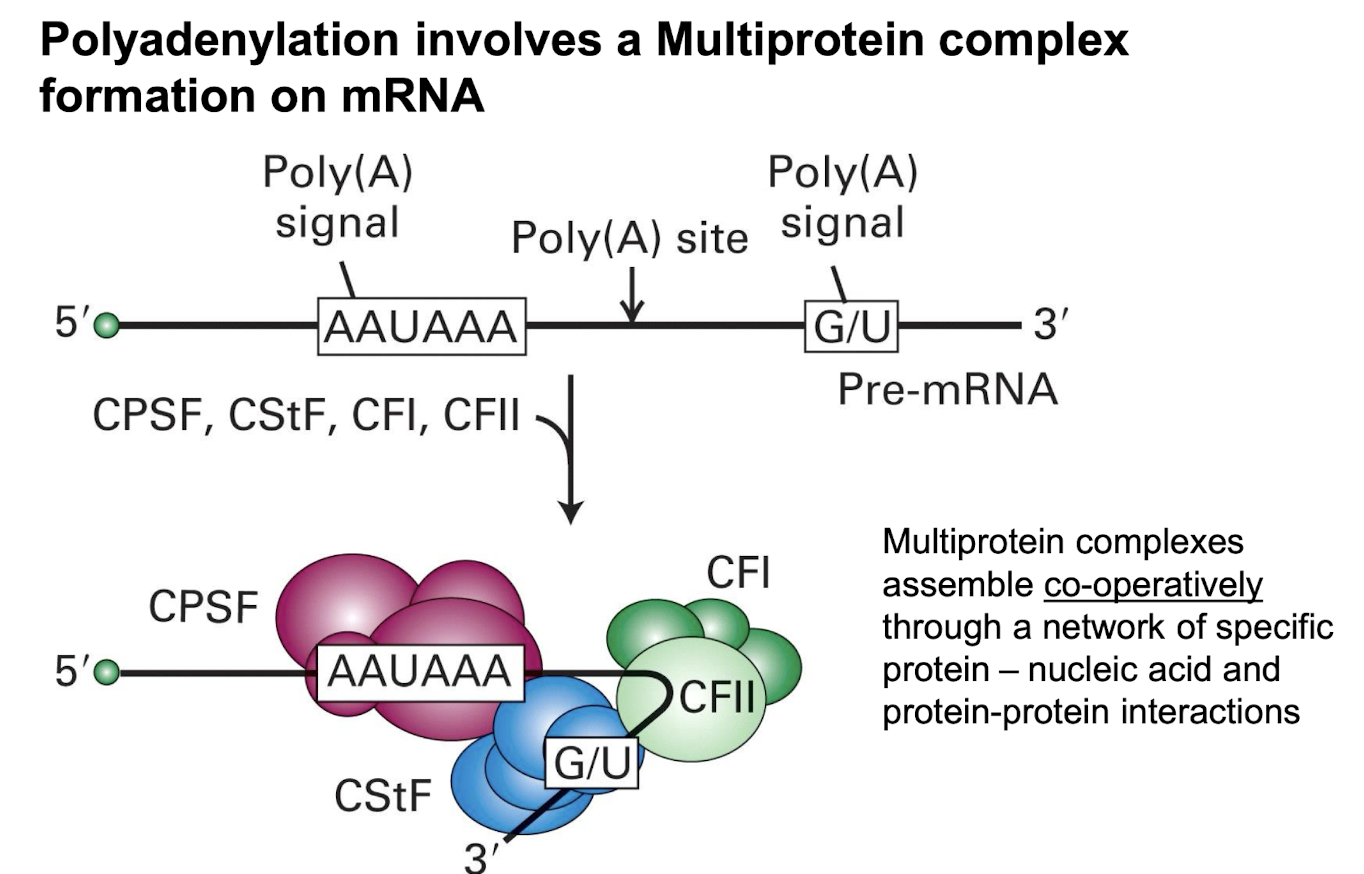
Polyadenylation involves formation of a _____
Multi-protein complex on mRNA which assembles operatively through network of specific protein-nucleic acid + protein-protein interactions:
CPSF = cleavage and polyadenylation specificity factor
CstF = cleavage stimulation factor F
CFI = Cleavage Factor I
CFII = Cleavage Factor II
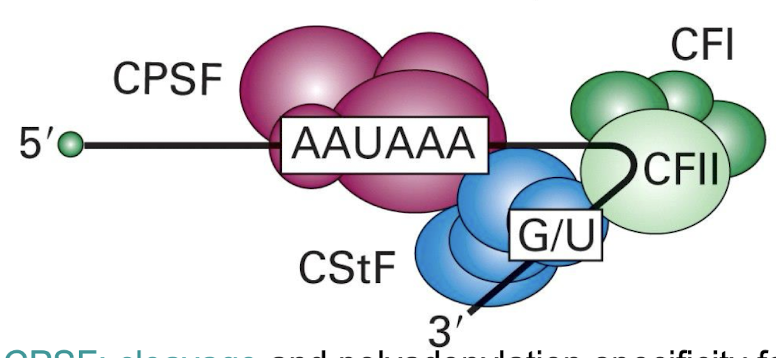
Function of CPSF and CstF?
Travel with RNA polymerase tail to the polyadenylation site, facilitating cleavage and poly(A) tail addition.
What needs to bind to the multi-protein complex before cleavage can occur?
Poly(A) polymerase (PAP)
Subsequent cleavage occurs exposing an OH group which initiates the addition of adenine residues to the 3' end of the mRNA.
What 2 phases of polyadenylation occur?
Slow polyadenylation
Rapid polyadenylation
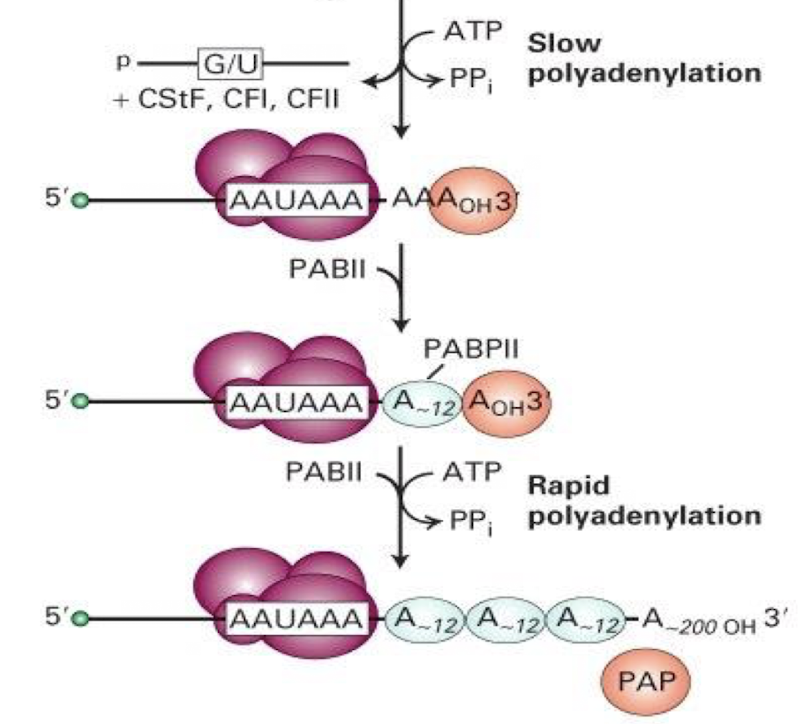
Slow polyadenylation involves?
the gradual addition of adenine residues to the 3' end of the pre-mRNA, mediated by poly(A) polymerase, leading to a brief pause before rapid completion.
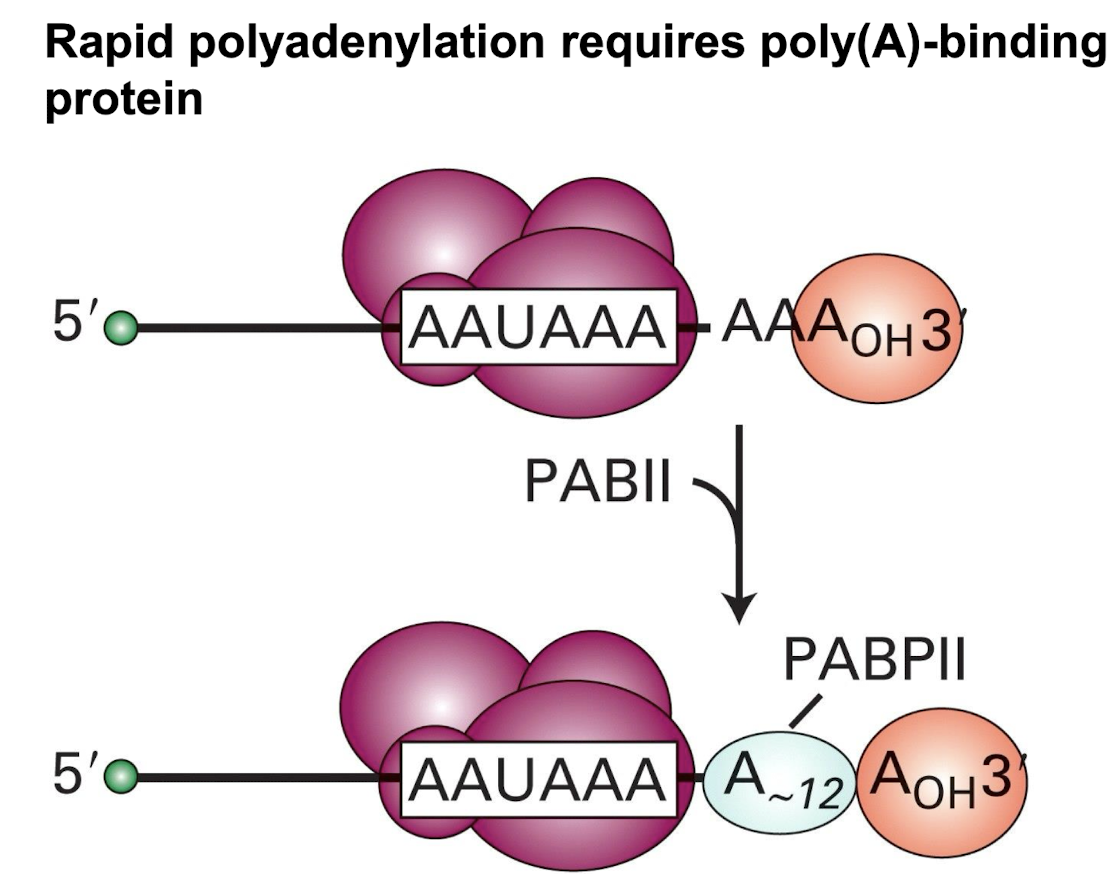
Rapid polyadenylation requires
PABII (Poly(A) binding protein II) to bind to the growing poly(A) tail and enhances the rate of adenine addition, up to 200-250 Adenosine residues addedto the 3' end of the mRNA.
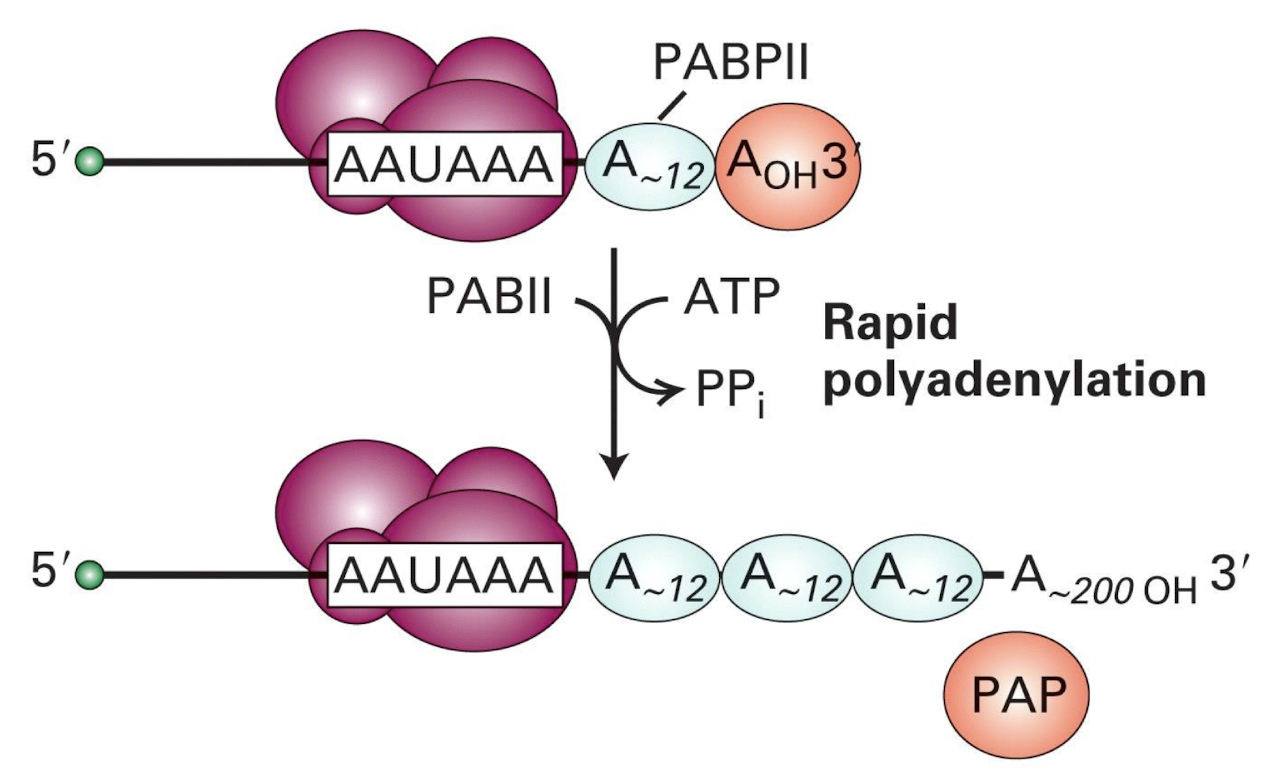
What are the 2 main functions of the poly A tail?
1) protects mRNA from degradation – PolyA signal (G/U) prevents exonuclease degradation of 3’ OH group
2) efficient translation - by facilitating ribosome binding and promoting mRNA stability, enhancing the overall translation efficiency.
What is the final step of forming a functional, mature mRNA?
RNA splicing, involving intron and exon removal which must occur at consensus seq around 5’ and 3’ splice sites at exon/intro boundry in pre-mRNAs
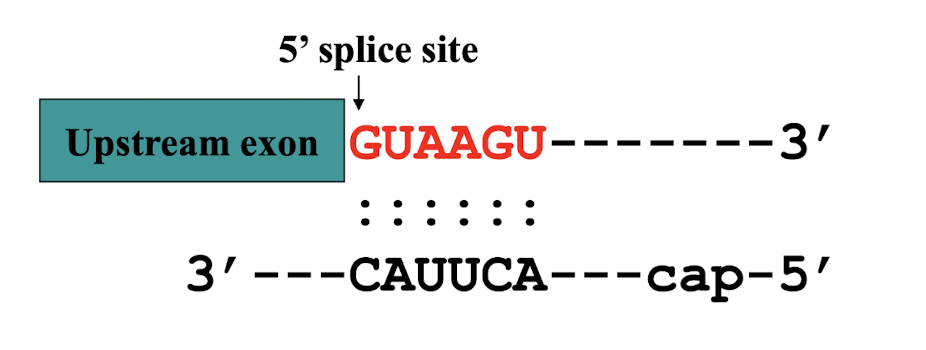
i.e. 5’ Splice site has conserved GU which is recognised by the splicing machinery
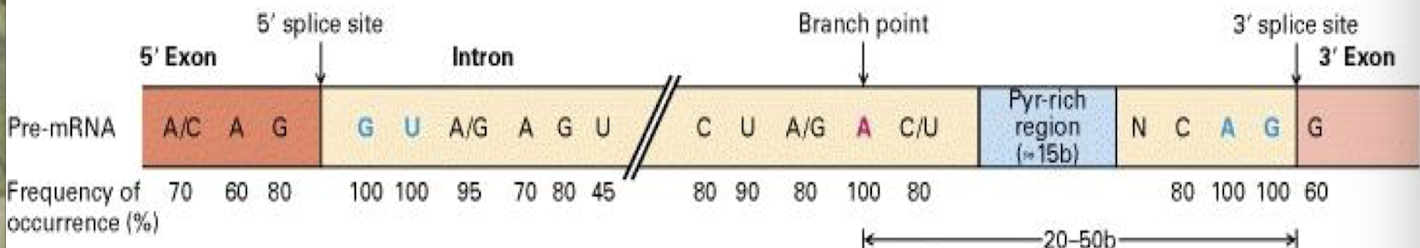
what do consensus sequences consist of?
▪ 5′ consensus sequence: GU A/G AGU: 5′ splice site
▪ 3′ consensus sequence: CAGG
▪ Branch point: the adenine “A”: ~18-40 nucleotides upstream of 3′-
splicing site
‒ Spliceosome: five RNA molecules + 300 proteins
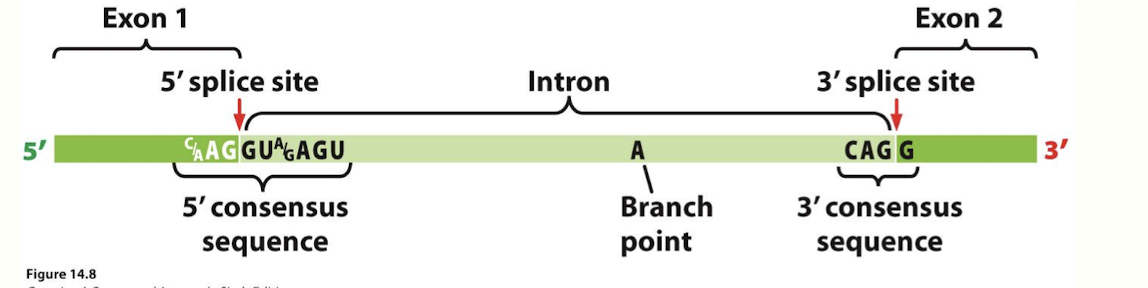
What are the 2 main RNA splicing reactions?
(2 sequential transesterifications)
Nucleophilic attack by the 2' (OH) hydroxyl group of the ADENINE in branch site on the 5' splice site leads to the formation of a lariat structure,
2nd nucleophilic attack by the 3' OH of exon 1 on the 3' splice site, attacks phosphodiester bond b/w intron 1 and exon 2 resulting in the ligation of exons and the release of the intervening intron.
*involves a SPLICEOSOME
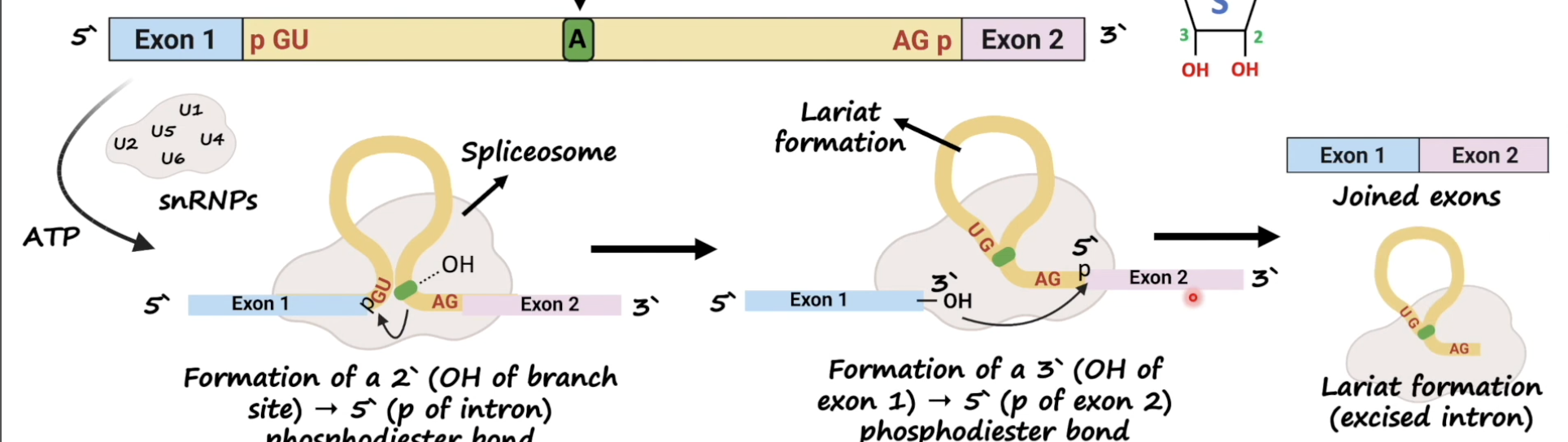
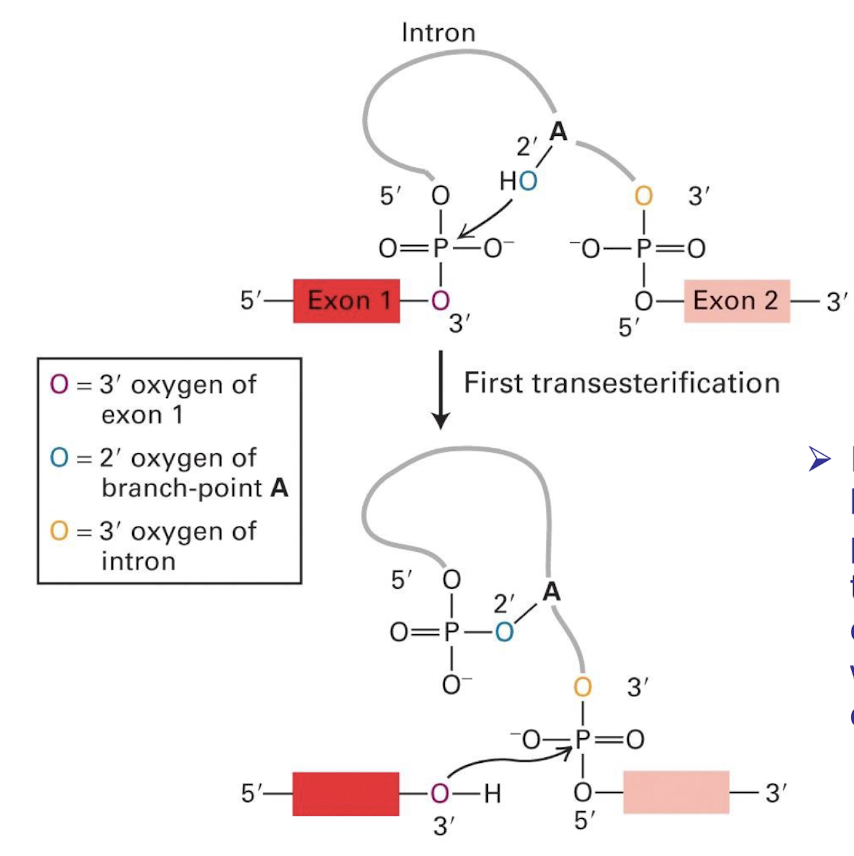
1st trans-esterification summarised
Nucleophilic attack by the 2' (OH) hydroxyl group of adenine in the branch site on the 5' splice site:
Ester bond between the 5′ phosphorus of the intron and the 3′ oxygen (red) of exon 1 is exchanged for an ester bond with the 2′ oxygen (dark blue) of the branch-site A residue
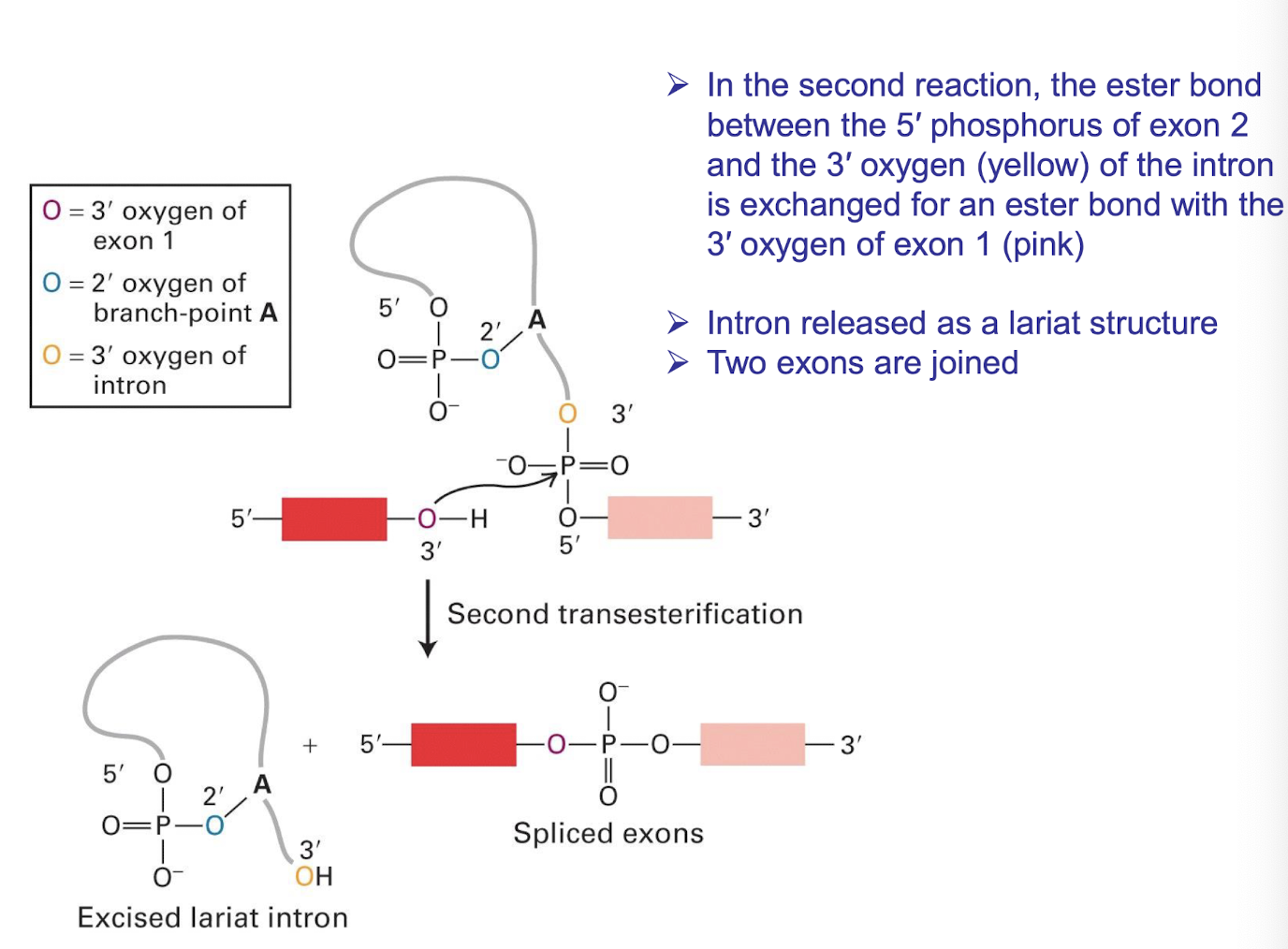
2nd trans-esterification summarised
The ester bond between the 5′ phosphorus of exon 2 and the 3′ oxygen (yellow) of the intron are exchanged for an ester bond with the 3′ oxygen of exon 1 (purple)
➢ Intron released as a lariat structure
➢ Two exons are ligated
what is the spliceosome and what is it composed of?
The spliceosome is a complex machinery responsible for the splicing of pre-mRNA.
Has Small Nuclear RNAs (snRNA) e.g. U1, U2, U5, U4/U6, each associating with >7 protein subunits
It is composed of small nuclear ribonucleoproteins (snRNPs) which form a large protein/ RNA complex
snRNA recognise intron-exon borders in primary RNA transcript
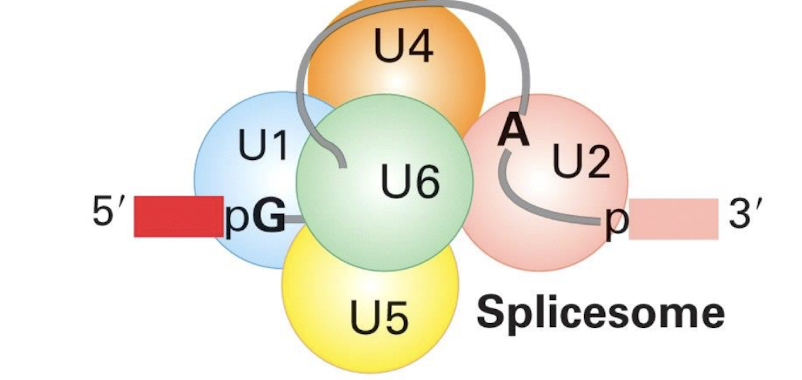
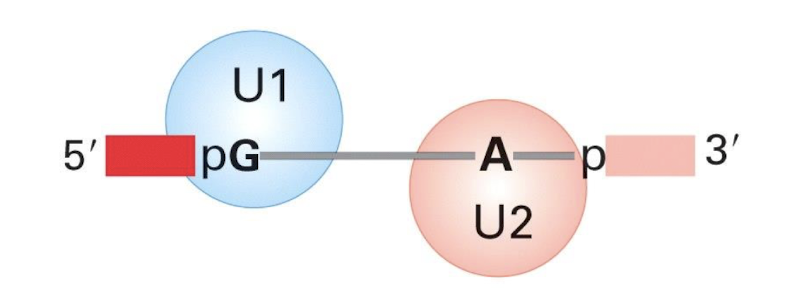
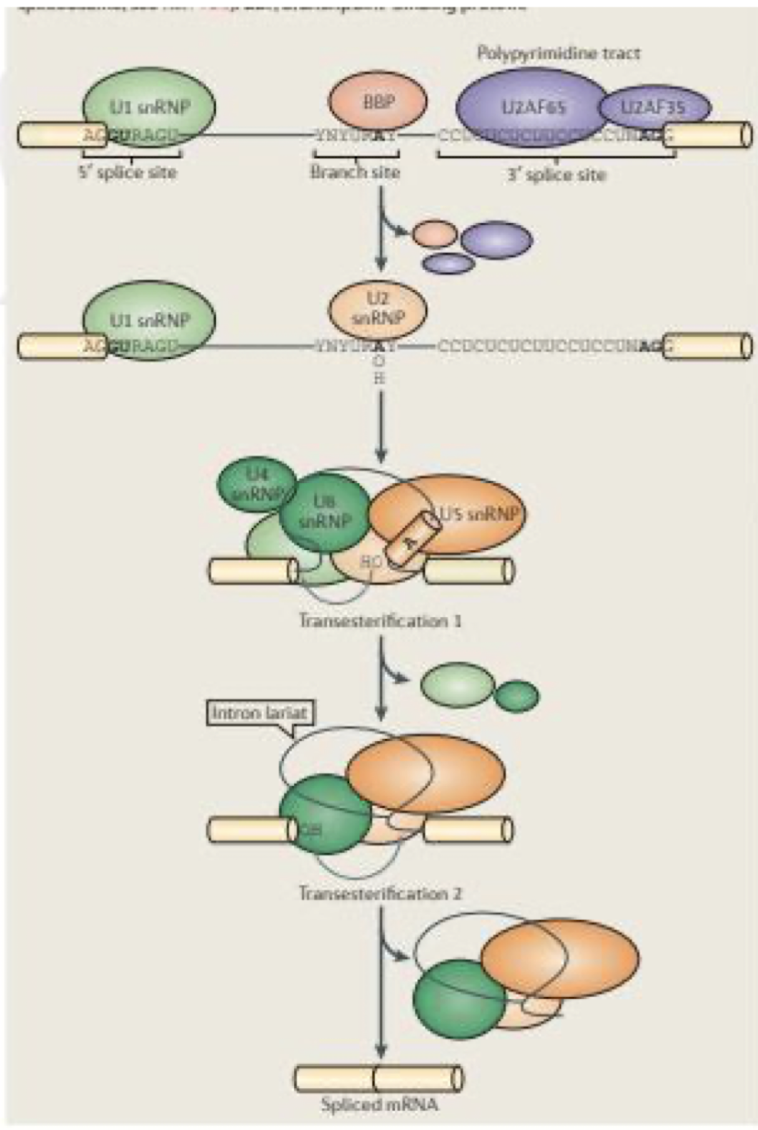
U1 binds to the …..
Binds 5' splice site and then the 3’ splice site
Contains conserved sequence complementary to 5’ splice site of nuclear mRNA introns
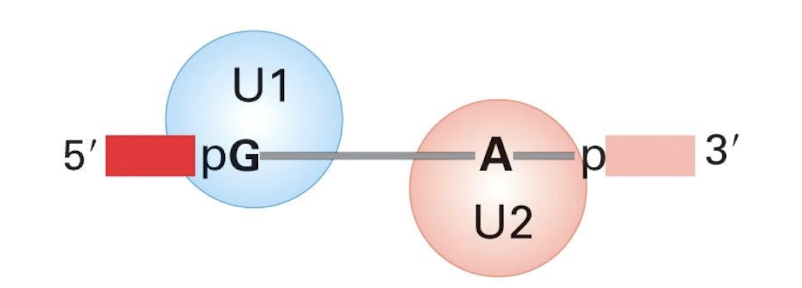
U2 binds the _________
Binds to the branch site (Adenine) and forms part of the catalytic centre with U6
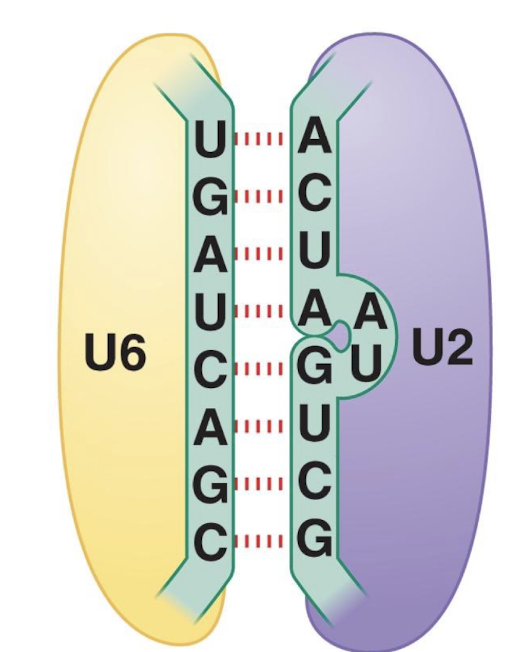
U4 masks
masks catalytic activity of U6
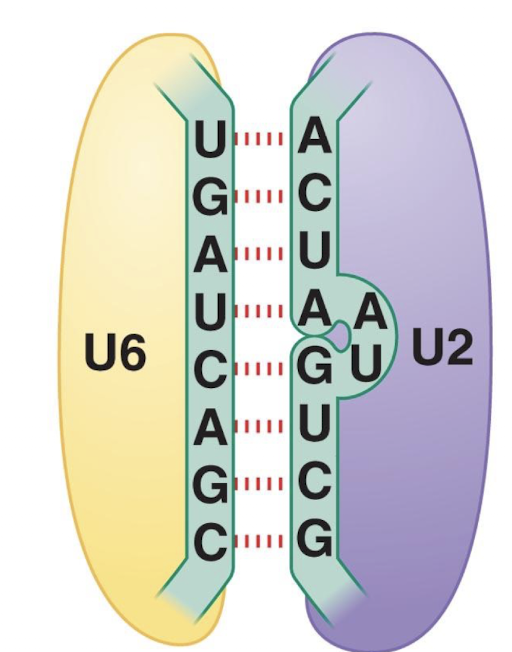
How does U4 act as an inhibitor of U6?
It masks U6 activity preventing pre-mature spliceosome assembly and once released allows U6 to interact with U2 (RNA:RNA interaction) forming the active site
Active site is primarily RNA and only present with spliceosome assembly
U6:U2 interaction positions substrate RNA
U5 binds to the
5’ splice site
U6 catalyses
the splicing reaction of pre-mRNA, facilitating the joining of exons and removal of introns.
Steps of spliceosome assembly:
U1 snRNA binds to 5’ exon - intron boundary
U2 binds, exposes nucleophilic adenine residue of branch site
U4/U6 complex forms and binds the pre-mRNA, allowing U5 to join and align the splice sites for catalysis - triple snRNP join complex
ATP-dependent unwinding disrupting base pairing b/w U4 + U6 → causes release of U1 and U4
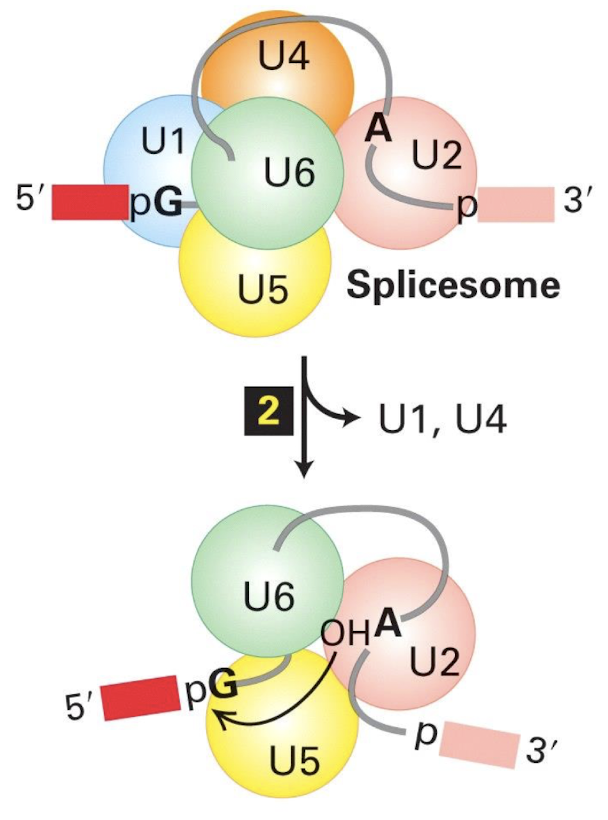
Nucleophilic attack by 2’ OH of branch point on 5’ Guanine of intron → 1st transesterification
Nucleophilic attack by 3’ OH of Exon 1 on 5’ G of exon 2 → 2nd 1st transesterification
Lariat formed and debranching enzyme forms linear intron RNA
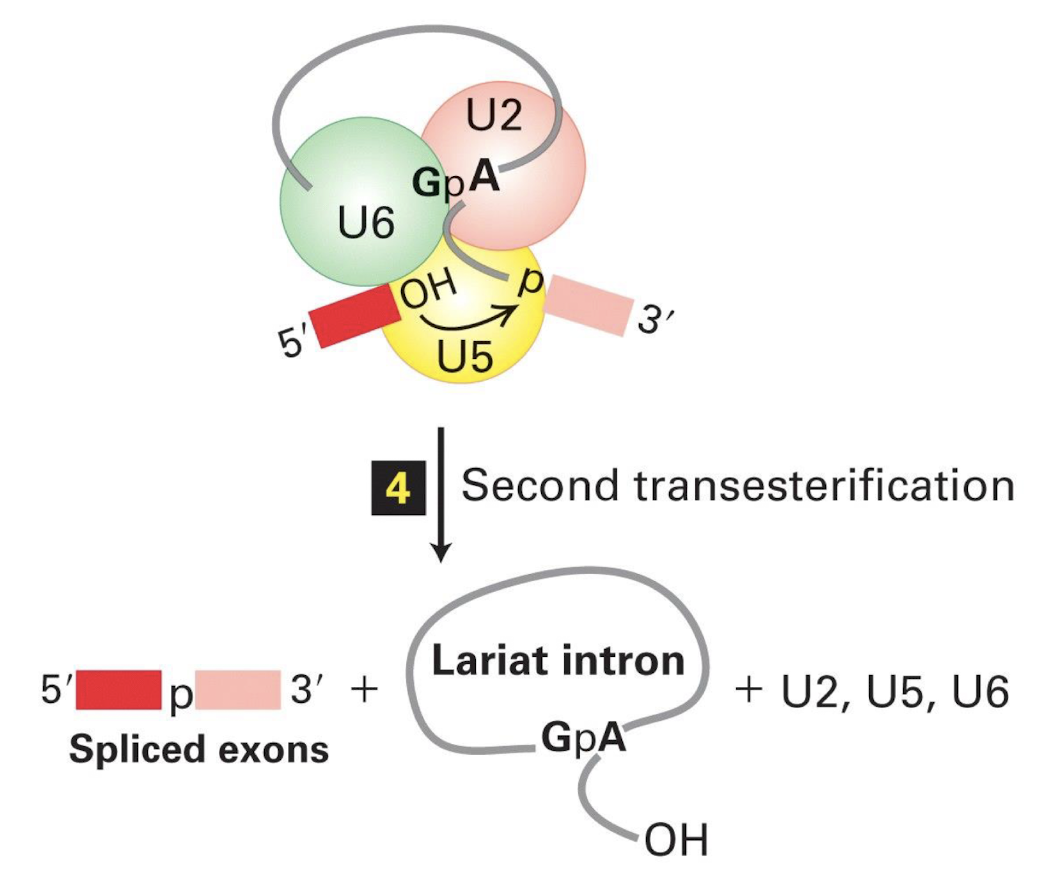
RNA splicing mechanism summarised
Spliceosome mediates a 2-step splicing reaction
The RNA component of the enzyme (snRNP) recognises the splicing signals on a pre-mRNA molecule, brings the two ends of the intron together, and provides the enzymatic activity for the two Transesterification reaction steps
Two Transesterification reactions do not require ATP
Activity of the spliceosome requires ATP hydrolysis
Summary (visual)
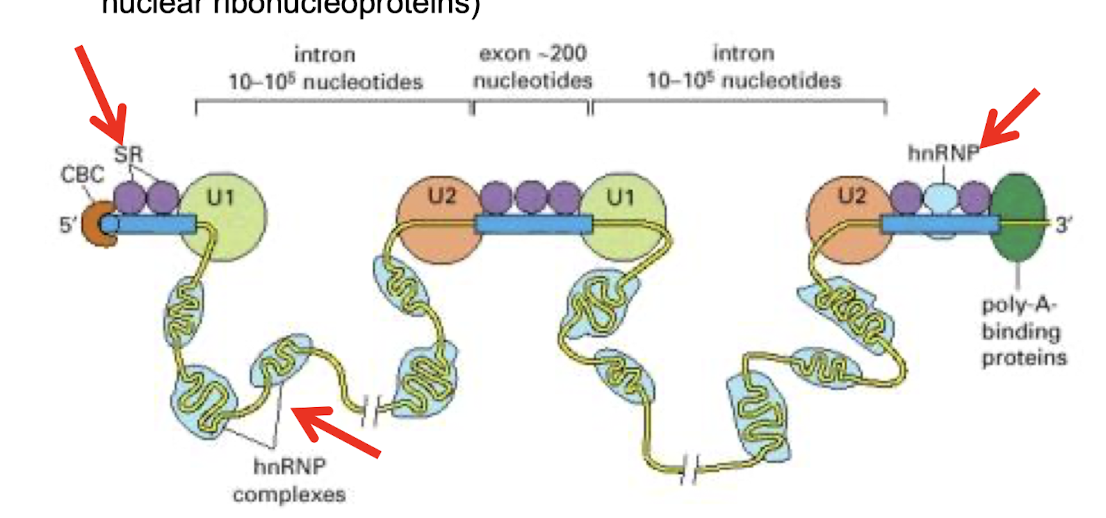
What are some points that reflect the fidelity of pre-mRNA splicing?
1. Spliceosome assembles as the pre-mRNA emerges from a transcribing RNA polymerase II
2. Exon definition hypothesis - spliceosome components, called the SR proteins (rich in Ser + Arg), assemble on exon sequences and mark each 3′ and 5′ splice site
3 Intron definition hypothesis – role of hnRNPs (heterogenous
nuclear ribonucleoproteins)
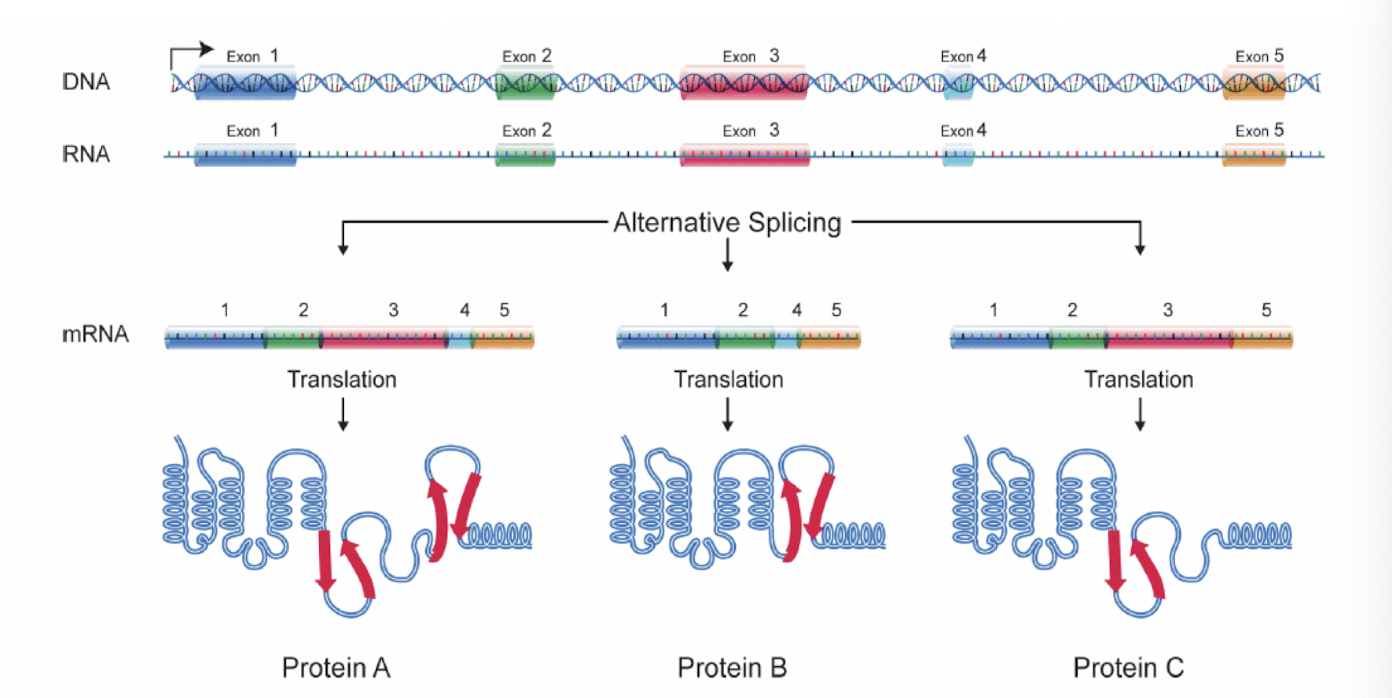
What is alternative splicing?
A process that allows a single gene to produce different mRNA and proteins by including or excluding specific exons during RNA splicing, which relates to the high complexity of transcriptomes in multicellular eukaryotes
Occurs in tissue specific + cell type specific manner, where deregulated AS can lead to diseases
What is an example of a well-studied AS gene?
AS of the Drosophila Down Syndrome Cell Adhesion Molecule (Dscam) gene.
Encodes an axon guidance receptor, similar to the human gene implicated in DS
Can generate over 38,016 different isoforms by extensive AS
The no. of proteins generated by this gene is 2-3X times the no. of genes in the entire Drosophila genome
AS of Dscam transcripts is regulated throughout development and in a tissue-specific manner
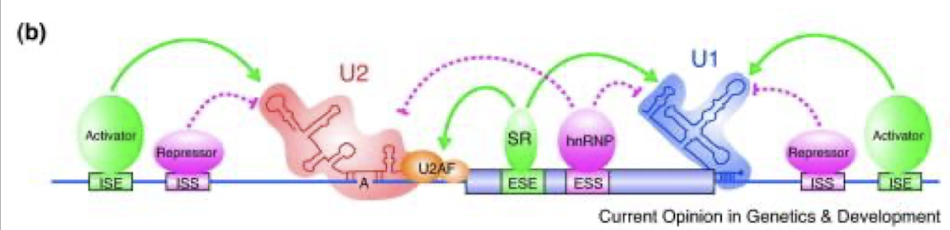
How is AS regulated by regulatory cis-sequences and trans-factors?
Alternative splicing (AS) regulation is influenced by various regulatory cis-sequences, such as splice sites and enhancers, along with trans-factors like splicing factors and regulatory proteins that interact with the spliceosome, ensuring the correct selection of splice sites and influencing mRNA diversity.
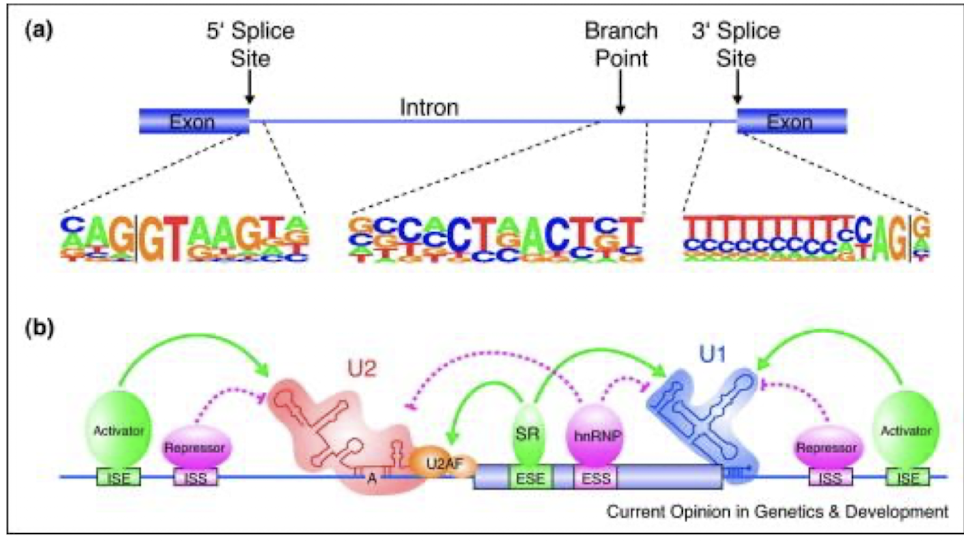
4 types of cis-regulatory RNA sequences bind splicing trans-factors
Intronic splicing enhancers (ISEs) - binds SR protein family
Intronic splicing silencers (ISSs) – binds Heterogeneous nuclear ribonucleoproteins (hnRNPs)
Exonic splicing enhancers (ESEs) - binds SR protein family
Exonic splicing silencers (ESSs) - binds hnRNPs
What are the main modes of AS of pre-mRNA?
Exon skipping
Mutually exclusive exons
Intron retention - rarer
Alternative 5' or 3' splice site selection
Alternative polyA site
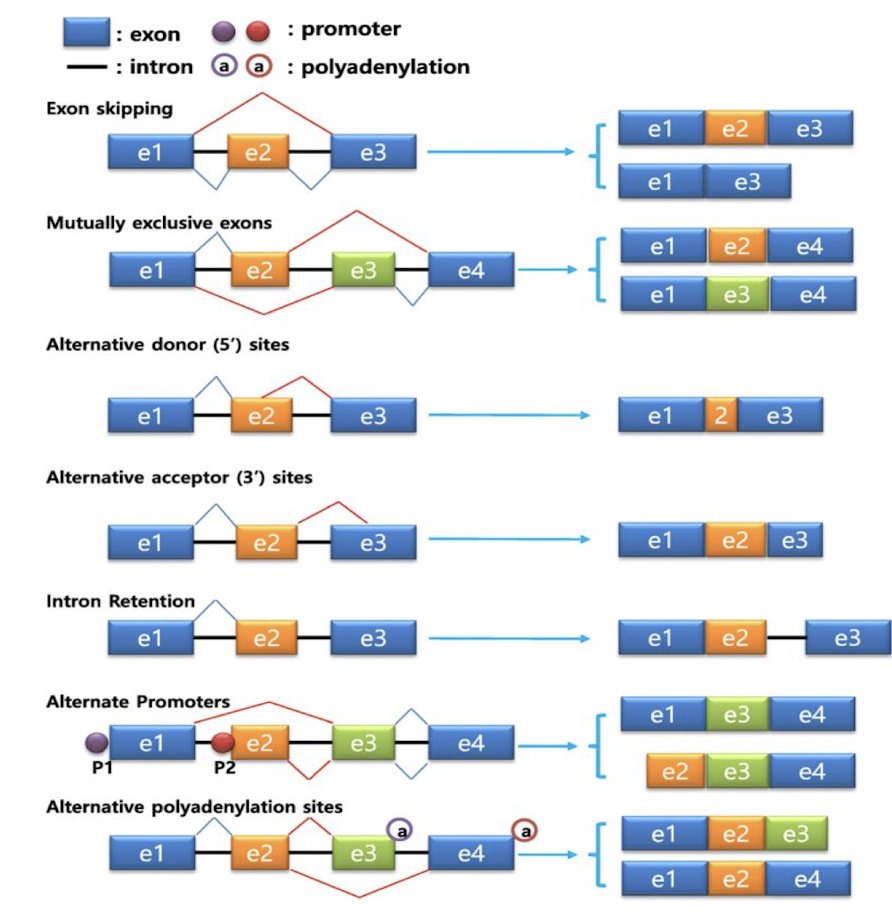
What is meant by steric hinderance in AS?
Where only one of two exons is removed but not both due to the physical obstruction caused by the presence of adjacent splice sites or regulatory elements, affecting the splicing process.
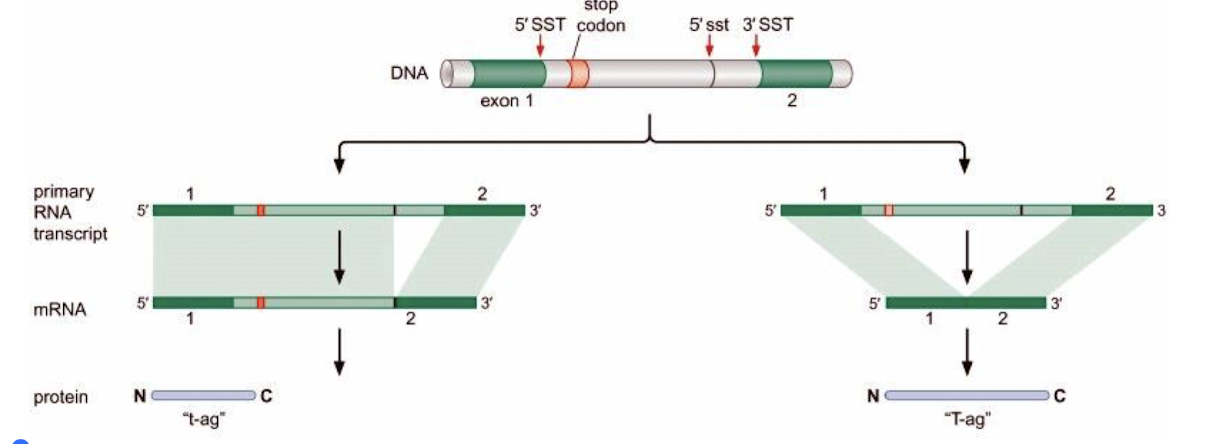
What is an example of what an alternative 5’ splice site can result in?
Constitutive alternative splicing results in 2 proteins: t-ag and T-ag, from the T antigen of the virus SV40
Different mature mRNAs result from the use of 2 different 5’ splice sites
In T (large), exon 1 is spliced to exon 2, deleting the intron
In t (small), the alternative 5’ splice site is used, resulting in the inclusion of a stop codon and shorter protein is produced
What is another example of what an alternative 5’ splice site can result in?
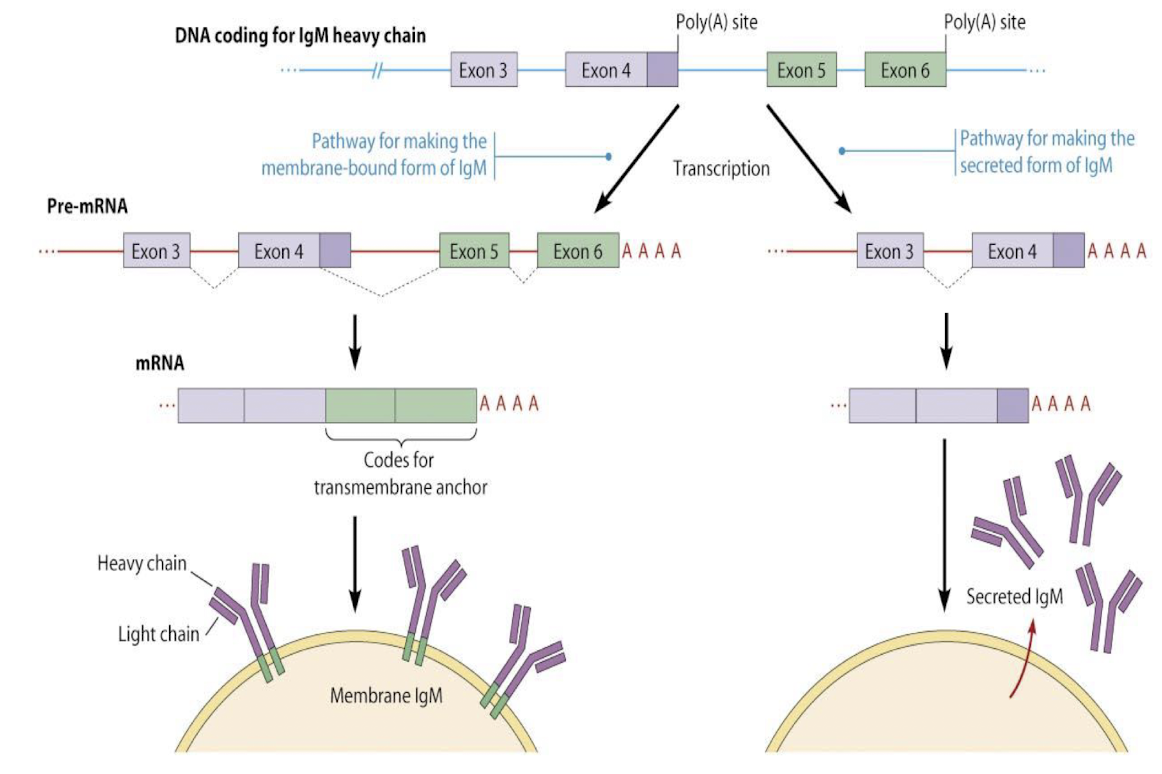
Alternative splicing of Antibody protein Immunoglobulin M (IgM) results in different protein forms: membrane-bound or secreted forms.
IgM produced determined by heavy chain C-terminus.
Alternative 5’ SS within Exon 4 controls use of PolyA site
This process allows for functional diversity in the immune response due to the use of different splice sites.
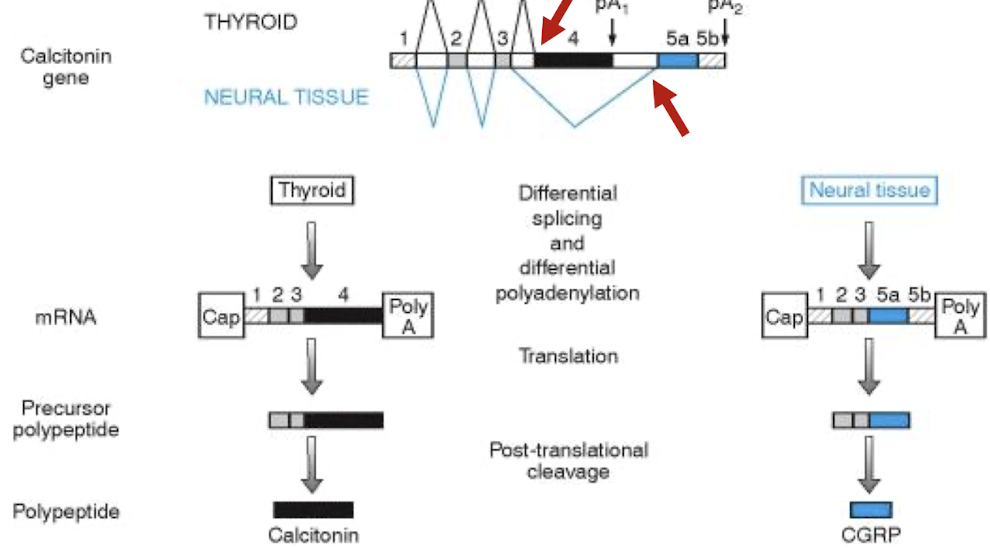
How is the tissue specific expression of calcitonin linked with AS and polyadenylation?
Calcitonin is encoded by Exon 4 sequences in the thyroid , while the calcitonin gene-related peptide (CGRP) synthesized in neural tissue, is encoded by the 5′ part of exon 5 (5a).
Controlled by alternative 3’ SS to control poly A signal
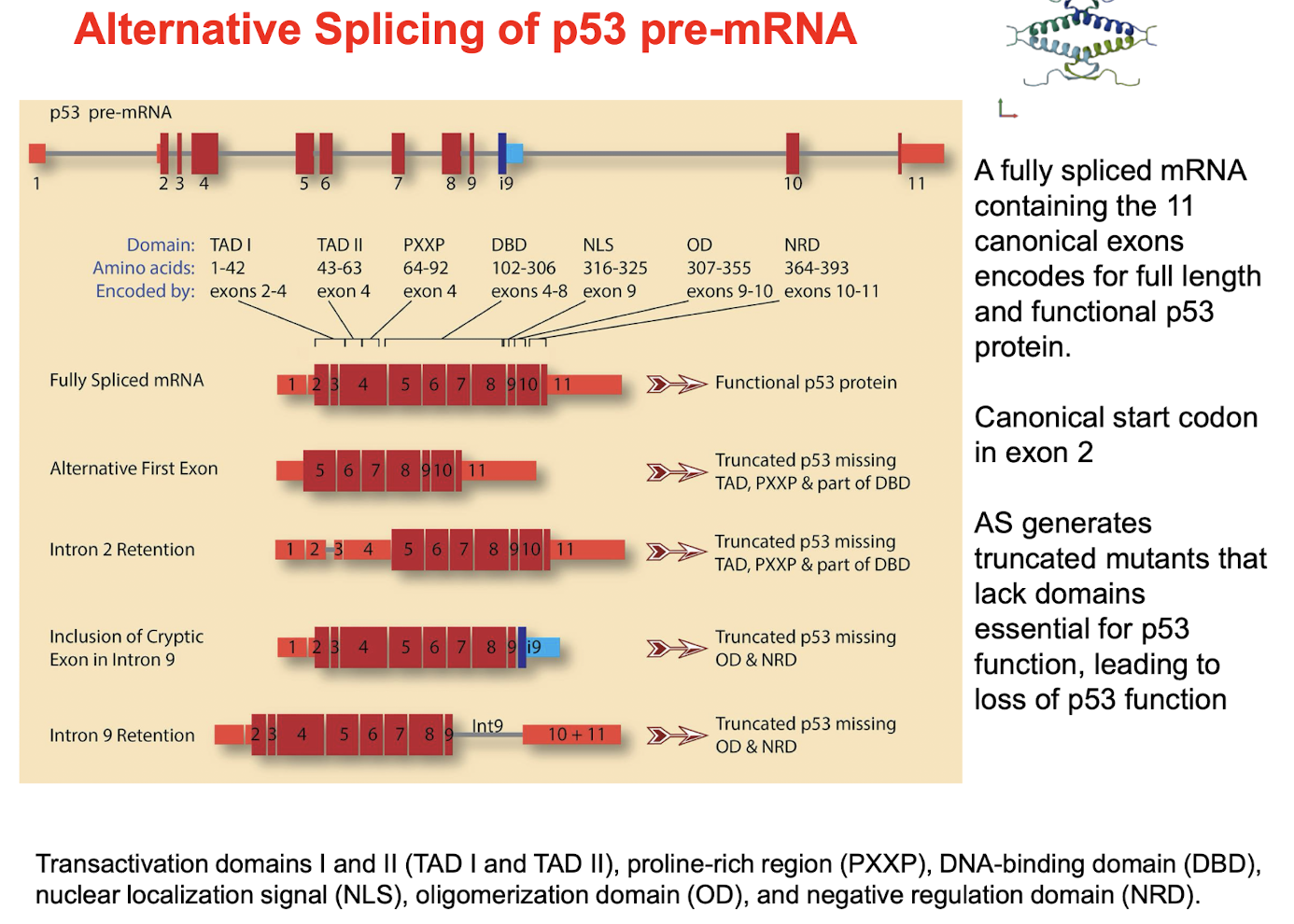
How is AS of p53 pre-mRNA linked with cancer?
Normally, A fully spliced mRNA containing the 11 canonical exons encodes for full length and functional p53 protein.
Canonical start codon in exon 2
AS generates truncated mutants that lack domains essential for p53 function, e.g. by inclusion / retention of intron 9 leads to loss of p53 function due to missing OD and NRD
What is RNA editing and an example?
Where the pre-mRNA sequence is altered by processes OTHER THAN RNA splicing
• Sequence of the corresponding mature mRNA differs from the exons encoding it in genomic DNA
•In higher eukaryotes, RNA editing is relatively rare, and only single-base changes have been observed.
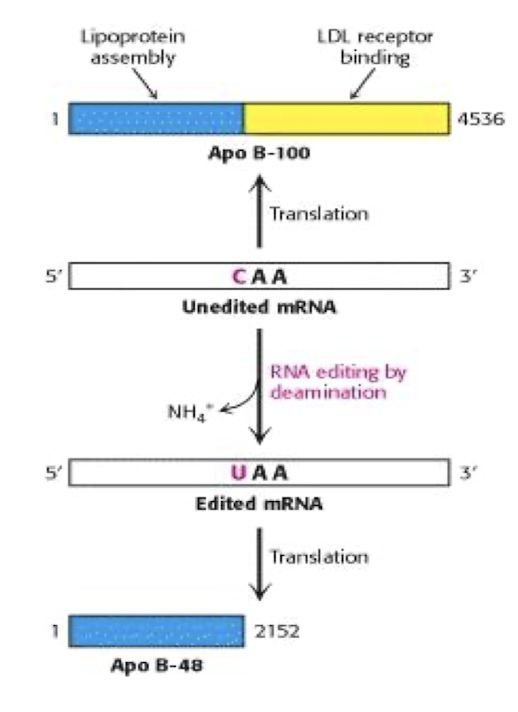
eg Apo-B, Glutamate receptor in Brain - important for triacylglycerol and cholesterol transport in by formation of an amphitatic spherical shell around lipids carried in lipoprotein particles
What 2 forms of apo B exist?
Apo B-100 synthesized by the liver, participates in cholesterol transport by binding LDL receptor
ApoB -48 -synthesized by the small intestine, carries dietary fat in the form of chylomicrons. Does not bind LDL receptor
2 functional domains of Apo-B100?
N-terminus associates with lipids
C-terminus binds LDL receptor (absent from Apo-B48) .
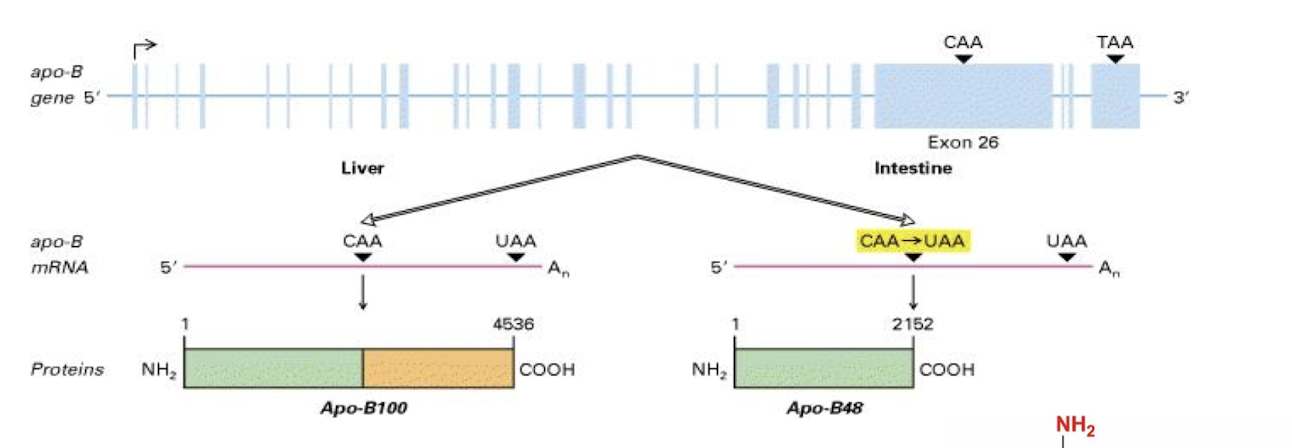
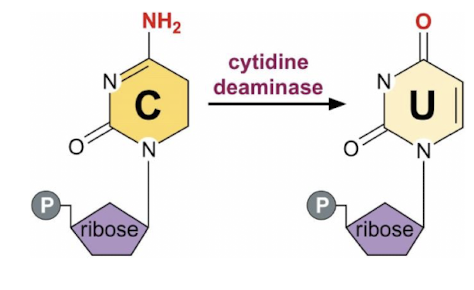
How is the RNA of apo-B edited?
The RNA of apo-B is edited by a process that changes a specific cytidine (C) to uridine (U) via cytidine deaminase found in the small intestine not liver & is expressed only at certain developmental stages,
resulting in the production of two different protein isoforms, Apo B-100 and Apo B-48, which have distinct functions in lipid metabolism