MLB 111 - Study Unit 5 Learning Outcomes
1/20
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
21 Terms
Explain how protein diversity is generated by only 20 amino acids.
The order of the amino acids and their individual chemical properties determine protein structure.
Recognize that amino acids differ in their biochemical properties, and that this is an important factor in protein structure and function.
They differ on chemical properties mostly conferred by the R-group. This includes hydrophobicity, charge, structure and reactivity.
Briefly describe the concept of primary, secondary, tertiary, and quaternary structure in proteins.
Primary - sequence of amino acids.
Secondary - interactions of nearby amino acids.
Tertiary - 3D shape of a polypeptide.
Quaternary - interactions of polypeptide subunits.
List the cellular components required for translation.
mRNA
Ribosomes
tRNA
Aminoacyl tRNA synthetases
Initiation factors
Elongation factors
Release factors
mRNA
Provides the template that specifies the amino acid sequence of the protein. It contains codons, which are sequences of three nucleotides that correspond to specific amino acids.
Ribosomes
Molecular machines that facilitate the translation process. They have two subunits (large and small) that come together during translation to form a functional ribosome. The ribosome reads the mRNA and catalyzes the formation of peptide bonds between amino acids.
tRNA
Brings the correct amino acids to the ribosome. Each tRNA has an anticodon that pairs with a specific mRNA codon and carries the corresponding amino acid.
Aminoacyl tRNA synthetases
Enzymes that charge tRNAs with the correct amino acids. Each aminoacyl-tRNA synthetase is specific for one amino acid and its corresponding tRNAs.
Initiation factors
Proteins required to start translation. They help in the assembly of the ribosome on the mRNA and the first tRNA. Examples include IF-1, IF-2, and IF-3 in prokaryotes, and eIFs (eukaryotic initiation factors) in eukaryotes.
Elongation factors
Proteins that facilitate the addition of amino acids to the growing polypeptide chain. Examples include EF-Tu and EF-G in prokaryotes and eEFs (eukaryotic elongation factors) in eukaryotes.
Release factors
Proteins that recognize stop codons and terminate translation. They help to release the newly synthesized polypeptide from the ribosome. Examples include RF-1 and RF-2 in prokaryotes and eRFs (eukaryotic release factors) in eukaryotes.
Make a simplified drawing of a ribosome.
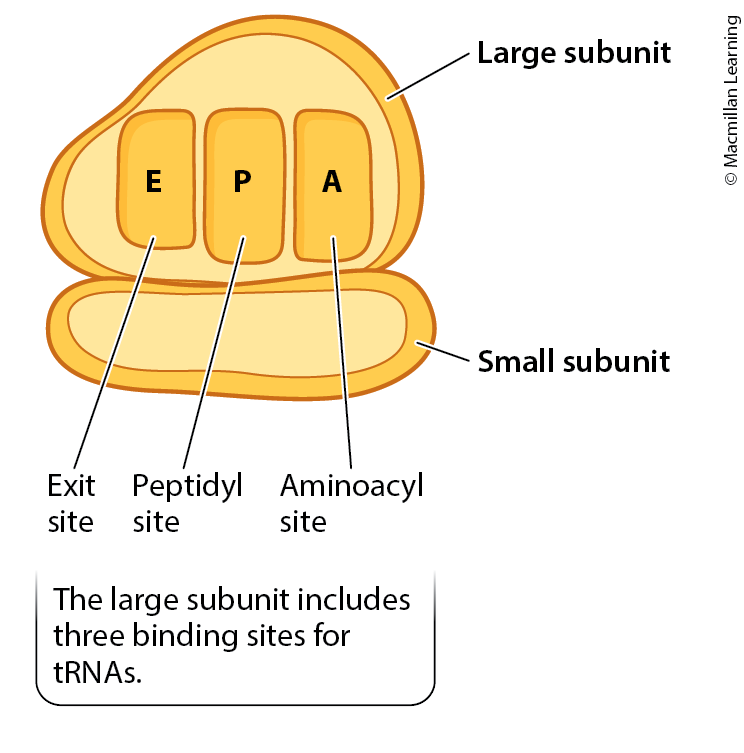
Explain the role of the reading frame, codon, anticodon and the process of base-pairing in the translation process.
Reading frame: The triplet (three base) grouping of RNA that is read uninterrupted. The reading frame that is used from an mRNA matters, and can produce different amino acid chains.
Codon: The triplet (three-base) code that specifies a specific tRNA and associated amino acid, or a release factor (for a stop codon).
Anticodon: The three-base sequence at the end of the tRNA that is complementary and anti-parallel to a specific codon.
Process of base-pairing: Base-pairing is the process by which nucleotides in the mRNA codon form hydrogen bonds with the complementary nucleotides in the tRNA anticodon.
Provide a simple labelled drawing of a transfer RNA.
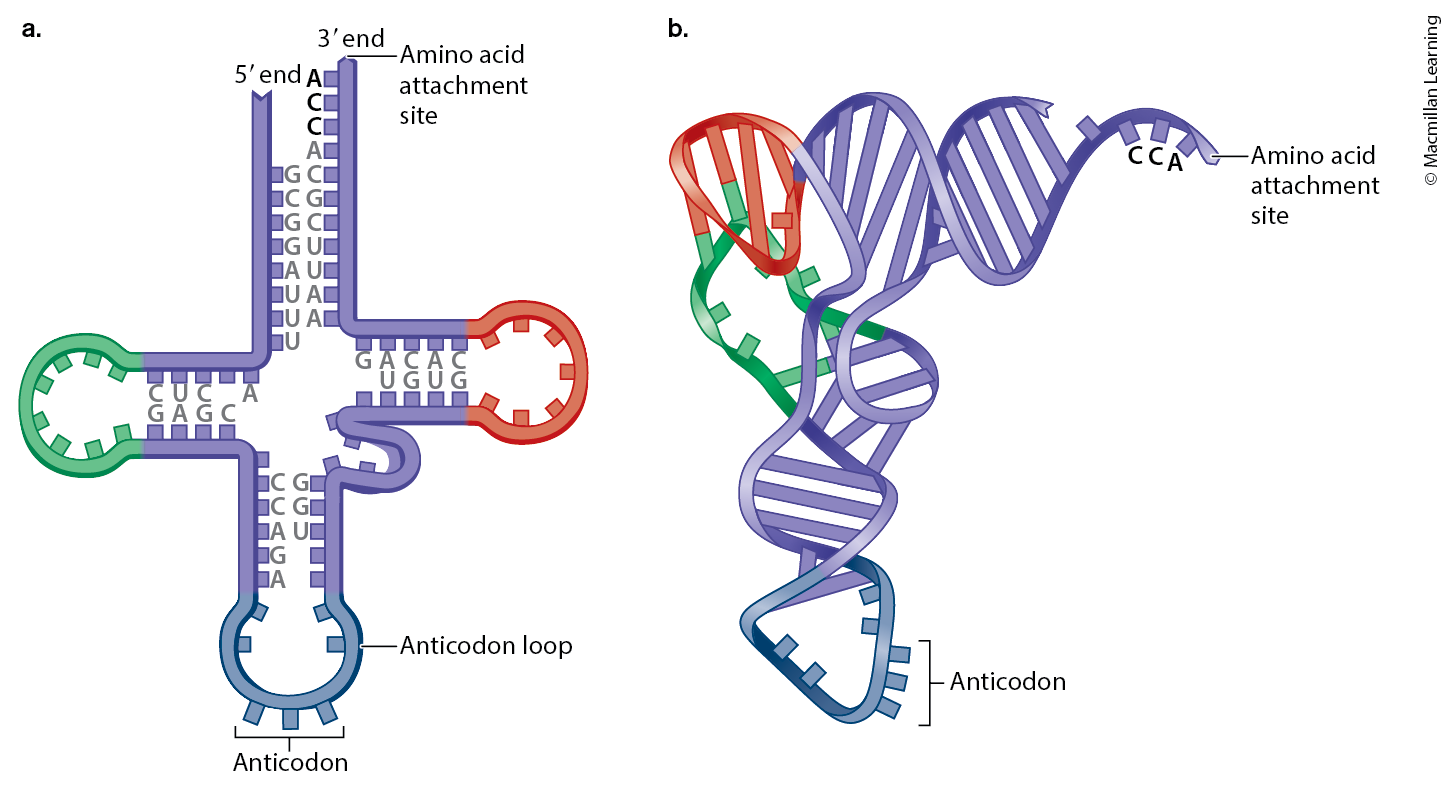
Explain the role of the aminoacyl tRNA synthetase in translation
Aminoacyl tRNA synthetase enzyme loads the appropriate amino acid to the correct tRNA:
One Aminoacyl tRNA synthetase per amino acid.
This is called charging the tRNA.
Provide an explanation of the genetic code and its role in translation.
The genetic code serves as the blueprint for translating the nucleotide sequence of mRNA into the amino acid sequence of proteins.
Ensures that the genetic information encoded in DNA is accurately and efficiently converted into functional proteins.
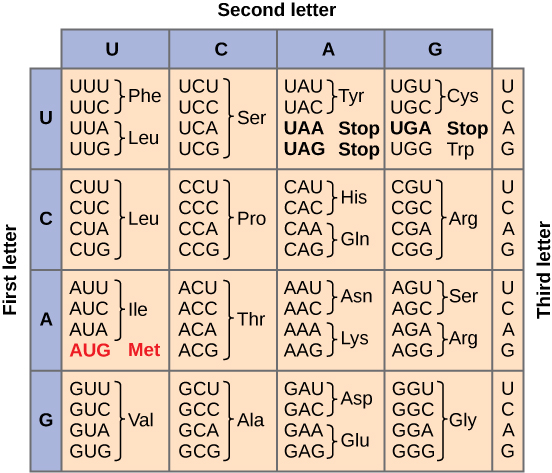
Provide a short description of the events that occur in each phase (initiation, elongation and termination) of translation.
Initiation: in this stage, the ribosome gets together with the mRNA and the first tRNA so translation can begin.
Elongation: in this stage, amino acids are brought to the ribosome by tRNAs and linked together to form a chain.
Termination: in the last stage, the finished polypeptide is released to go and do its job in the cell.
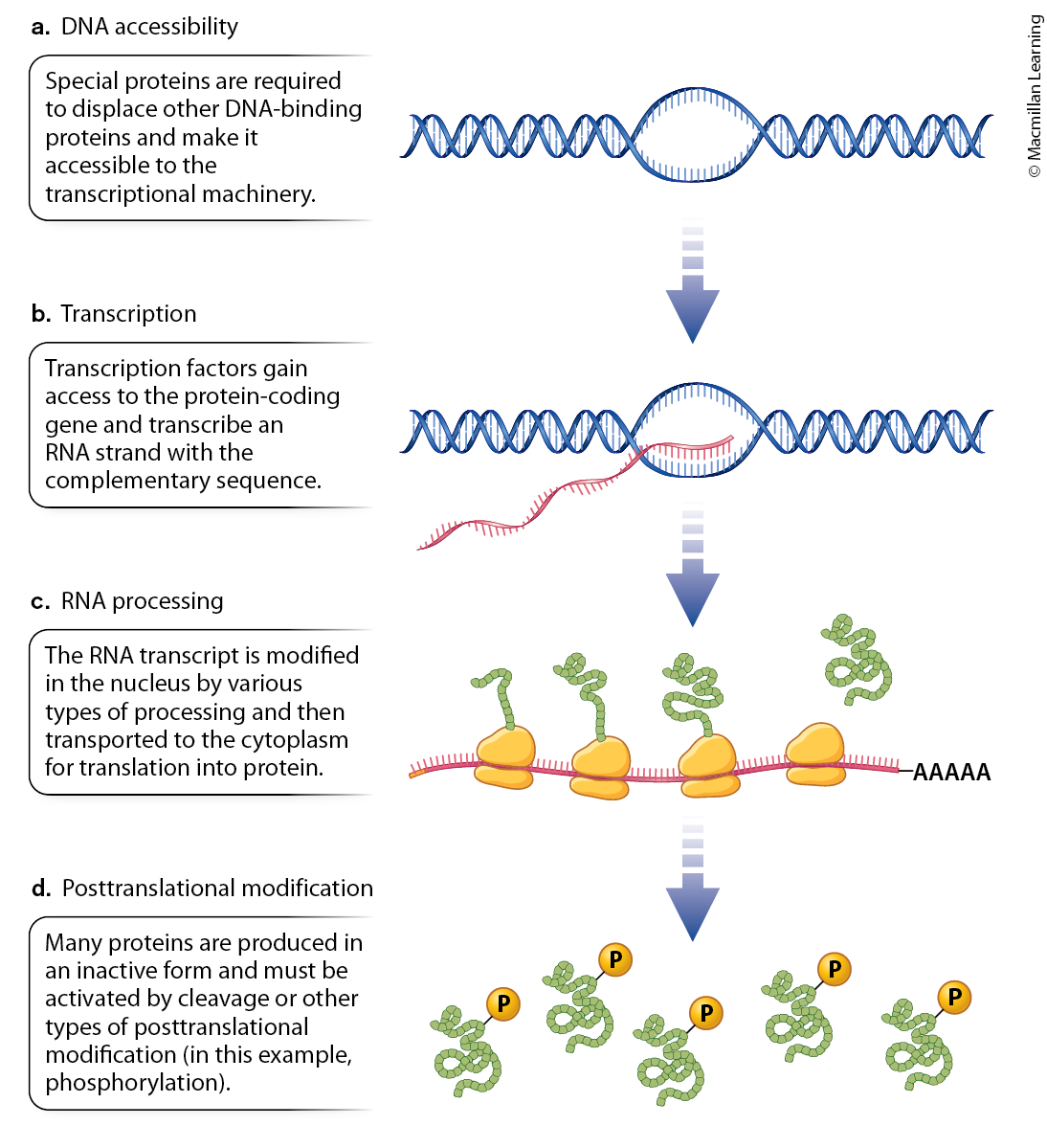
How do we regulate gene expression?
DNA needs to be accessible; Ensures that genes are in an open chromatin state to be transcribed.
Transcription factors need to drive transcription; Drive the initiation and regulation of transcription by binding to specific DNA sequences.
RNA processing through splicing, capping and adding a poly-A tail; Modifies pre-mRNA through splicing, capping, and polyadenylation to produce mature, translatable mRNA.
Modification of the protein to influence its function; Alters the protein’s function, stability, and localization after it is synthesized.
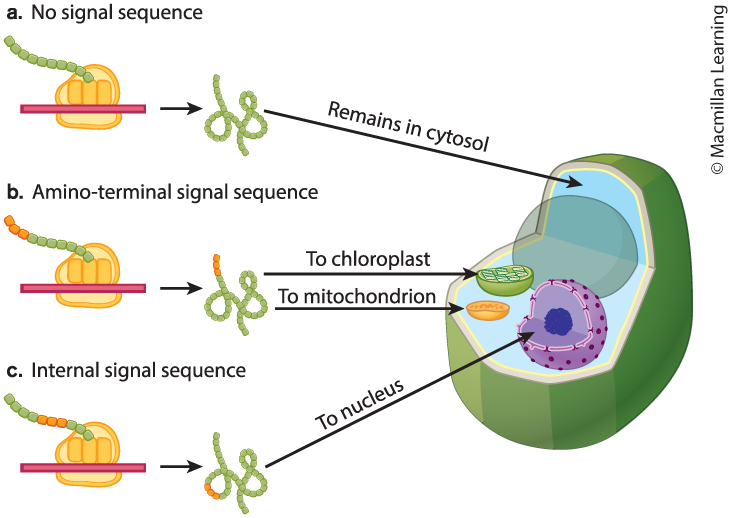
Where do proteins go? [Protein Sorting]
Protein sorting directs proteins to their final destinations inside or outside of the cell.
Stays in the cytosol.
Transferred to the organelles.
Transferred to the nucleus.
Incorporated into a membrane.
Briefly explain the mechanisms that affects protein sorting, by referring to the destination of proteins synthesized on free ribosomes versus those synthesized on the rough ER.
Proteins synthesized on free ribosomes are sorted after translation, whereas proteins synthesized on ribosomes associated with the rough ER are sorted during translation.
Proteins synthesized on free ribosomes are often sorted by means of a signal sequence and are targeted for the cytosol, mitochondria, chloroplasts, or nucleus.
Proteins synthesized on ribosomes on the rough ER have a signal sequence that is recognized by a signal-recognition particle. These proteins end up as transmembrane proteins, reside in the interior of various organelles, or are secreted.
Provide a brief description of how mutations and subsequent natural selection drive the evolution of novel proteins. Include the limitation on protein folding as part of your discussion.
Proteins evolve by combining different folding domains (A region of a protein that folds in a particular way and that carries out a specific function).
Proteins also evolve through changes in their amino acid sequence, which occurs by mutation and natural selection.
The probability that a random sequence of amino acids will fold properly to carry out a specific function is very small.