The Eukaryotic Cell Cycle Chapter 19
1/94
Earn XP
Description and Tags
Bolded words from slides defined
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
95 Terms
Mitosis
Cell division that generates daughter cells genetically identical to their parent. The basis of growth, replacement, and regeneration.
In eukaryotic cells, the process whereby the nucleus divides, producing two genetically equivalent daughter nuclei with the diploid number of chromosomes.
Meiosis
Reductive division that halves the number of chromosomes in the resulting cells (gametes) which also recombining genetic information from maternal and paternal chromosomes to generate genetic diversity
In eukaryotes, a special type of cell division that occurs during maturation of germ cells; comprises two successive nuclear and cellular divisions with only one round of DNA replication. Results in production of four genetically nonequivalent haploid cells (gametes) from an initial diploid cell.
Sister Chromatid
The two identical DNA molecules created during DNA replication and the associated chromosomal proteins. After DNA replication, each chromosome is composed of two sister chromatids.
Sister chromatid resolution
The process of untangling of the intertwined sister chromatids during prophase.
Mitotic Cell Cycle
Orderly sequence including:
G Phase
S Phase
M Phase
G Phase
Cell growth and preparation for the next stage of cell cycle
G1: Cell growth, normal cell activities AND monitoring environment for signals to initiate division
G2: More growth and inspection to ensure faithful chromosome replication- diploid organisms at this point have a 4n complement of chromosomes
S Phase
Faithfully copy all chromosome forming two identical sister chromatids
DNA synthesis- each parental chromosome is copied into identical sister chromatids.
M Phase
Faithfully segregate sister chromatids and other cellular material to daughter cells
following M phase and cytokinesis each daughter cell contains a 2n complement of chromosome
Cell Cycle
Ordered sequence of events in which a eukaryotic cell duplicates its chromosomes and divides into two. The cell cycle normally consists of four phases: G1 before DNA synthesis occurs; S when DNA synthesis occurs; G2 after DNA synthesis; and M when cell division occurs, yielding two daughter cells. Under certain conditions, cells exit the cell cycle during G1 and remain in the G0 state as non-dividing cells.
Tightly monitored through each of these phases. Entry/exit of each phase is tightly regulated to ensure fidelity of both replication and segregation.
Checkpoint Pathway
Surveillance mechanism that prevents initiation of each step in cell division until earlier steps on which it depends have been completed and mistakes that occurred during the process have been corrected.
monitor fidelity of each stage and prevent progression into the next until all appropriate tasks are complete.
Cell Cycle Checkpoint Pathways
Prevent initiation of the next stage of the cycle until the earlier step has been completed, and any mistakes have been corrected.
Cytokinesis
The division of the cytoplasm following mitosis to generate two daughter cells, each with a nucleus and cytoplasmic organelles.
Interphase
G1, S, and G2 are collectively called interphase.
G1 Checkpoint
Are conditions right to initiate division? Unicellular organisms, like yeast, need critical nutrients; in multicellular organisms, appropriate signals and contact to the extracellular matrix is necessary.
This checkpoint is called START in yeast and the Restriction Point in mammals. if the cell passes through the restriction point it is irreversibly committed to preparing for and completing DNA replication.
G2 Checkpoint
Have all chromosomes been replicated and have any errors been corrected?
Metaphase Checkpoint
Aka: Spindle assembly checkpoint
Are all replicated chromosomes at the metaphase plate and have all sister chromatids been attached to microtubules.
Prevents progression into anaphase unit all chromosomes are bioriented
Sensor proteins
monitor these conditions and relay information back to the proteins that drive the transition from one stage of the cell cycle to the next.
G1 cyclin-CDKs
Cyclin-CDK complexes that promote entry into the cell cycle
Entry into cell cycle. Its activity is regulated by signal transduction pathways that respond to growth factors or anti-proliferation signals. For example, they are activated downstream of the Ras/MAPK pathway.
G1/S phase cyclin CDKs
Cyclin-CDK complexes that promote entry into the cell cycle together with G1 CDKs.
Cyclin levels accumulate in late G1 and trigger the G1 to S phase transition
the mammalian restriction point
Mitotic cyclin- CDKs
Cyclin-CDK complexes that promote entry into and progression through mitosis.
The cyclins are synthesized during S phase but their activity is held in check until DNA synthesis is complete.
S-phase cyclin-CDKs
Cyclin-CDK complexes that promote the initiation of DNA replication
Promote DNA replication by activating DNA helicase and loading DNA polymerase onto chromosomes.
G0
Not all cells divide
Most terminally differentiated cells exit the cell cycle at G1 and enter an arrested state called G0
Quiescence
Some cells are only temporarily “arrested” can be induced to become mitotically active again.
Senescence
Cell have lost the ability to receive signals (usually due to age) that would initiate cell division and actively repress genes needed for mitosis.
Cohesin
Protein complex that holds the replicated sister chromatids together.
Join sister chromatids along their lengths.
Prophase
Earliest stage in mitosis, during which the chromosomes condense, the duplicated centrosomes separate to become the spindle poles, and the mitotic spindle begins to form.
Duplicated chromosomes condense
Centrosomes begin migrating to opposite poles of cell, radiating microtubules to generate the mitotic spindle
Cohesin is removed from chromosome arms, but sister chromatids remain connected by cohesion at the centromere
Kinetochore assembles on centromere
Centromere
DNA sequence required for proper segregation of chromosomes during mitosis and meiosis; the region of mitotic chromosomes where the kinetochore forms and that appears constricted.
Kinetochore
A multilayer protein structure at or near the centromere of each mitotic chromosome form which microtubules extend toward the spindle poles of the cell; plays an active role in movement of chromosomes toward the poles during anaphase.
Kinetochore-associated tension sensing mechanism aligns sister chromatids at the metaphase plate.
Prometaphase
Second stage in mitosis, during which the nuclear envelope and nuclear lamina break down and microtubules assembled from the spindle poles “capture” chromosome pairs at specialized structures call kinetochores.
Chromosomes continue to condense
Nuclear envelope breaks down and retracts into ER
microtubule (+) ends find and attach to kinetochores
Metaphase
Stage of mitosis at which condensed chromosomes are aligned equidistant between the poles of the mitotic spindle but have not yet started to segregate toward the spindle poles.
Each sister chromatid is attached to microtubules originating from opposite poles
Chromosomes align at central region called the metaphase plate
Anaphase
Mitotic stage during which the sister chromatids (or duplicated homologs in meiosis I) separate and move apart (segregate) toward the spindle poles.
Microtubule shortening pulls separated chromatids to opposite poles
Other microtubules lengthen, and aided by kinesin motor proteins, the spindle, and cell elongate
Telophase
Final mitotic stage, during which the nuclear envelope re-forms around the two sets of separated chromosomes, the chromosomes decondense, and division of the cytoplasm (cytokinesis) is completed.
Chromosomes arrive at opposite pole-begin to decondense
Nuclear envelope begins to reassemble
Mitotic spindle begins to disassemble
Actin-based contractile ring (animal cell specific) begins to assemble at midpoint.
Cytokinesis
In animal cells a cleavage furrow forms resulting from actinomyosin contraction of the contractile ring. In plant cells, the cell plate- precursor of the new cell wall forms to separate the new daughter cells.
The division of the cytoplasm following mitosis to generate two daughter cells, each with a nucleus and cytoplasmic organelles.
Mitotic spindle
A specialized temporary structure, present in eukaryotic cells during mitosis, that catures the chromosomes and then pushes and pulls them to opposite sides of the dividing cell; also called mitotic apparatus.
Anaphase A and B
Distinct mechanisms aggregate duplicated chromosomes to opposite poles and push the poles apart, lengthening the cell in preparation for division.
Centrosome cycle
During S phase, animal cells duplicate their centrosome in coordination with chromosome replication.
paired centrioles separate- each buds a new daughter centriole
G2- daughter centriole duplication is complete, but the two pairs of centrioles remain within a single centrosome
At the G2 and M phase transition, the centrioles separate and migrate to opposite poles of the nucleus
Aster
Structure composed of microtubules (astral fibers) that radiate outward from a centrosome during mitosis.
aka spindle poles
Astral MTs
Project toward and engage to the cell cortex (microfilament web beneath the plasma membrane). This anchors centrosomes in place and orients the spindle to generate the axis of division.
Polar MTs
Project toward the cell center. These microtubules fail to capture chromosomes. Instead, they become aligned antiparallel to polar MTs from the opposite pole. During anaphase this will drive 1) separation of poles and contribute to chromosome separation 2) and cell elongation
Kinetochore MTs
Capture sister chromatid in replicated chromosomes and will also contribute to chromosome separation during anaphase.
CENPA
special 3 histone protein which DNA is bound to
Ran in mitosis
A GEF for the nuclear import factor Ran is bound to chromosomes during mitosis
generate high concentration of Ran-GTP around the centromere
Ran-GTP releases a protein from importin called TPX which binds augmin and the ᵞTURK complex. This creates very shallow branches from chromosomes approaching chromosomes and enhances the number of microtubules that bind chromosomes.
Bioriented
When both kinetochores of a replicated chromosome are bound by microtubules.
Indicates that the kinetochores of sister chromatids have attached to microtubules emanating from opposite spindle poles.
Dynein-Dynactin
Complexes on both kinetochores pull each sister chromatid toward their respective poles, bioriented chromosomes experience tension.
Congression
chromosome movement toward the cell center- the metaphase plate.
Kinesin-4
Another process contributing to congression
on chromosome tips move on polar microtubules toward their (+) ends- this pulls chromosomes toward the cell center.
While this happens, a kinetochore tension sensing mechanism reads which direction the chromosomes are being pulled and
promotes polymerization of microtubules on the short side, growing microtubules are tethered to kinetochore by kinesin-7
While kinesin-13 on the other kinetochore promotes microtubule depolymerization
cells will not progress from metaphase to anaphase unitl all chromosomes are bioriented
Ndc80
Microtubules are captured by this outer kinetochore protein complex, its like a sleeve that a microtubule can slide into.
Aurora kinases
Serine/threonine kinases that play a crucial role in cell division by controlling chromatid segregation. Aurora B kinase destabilizes faulty microtubule-kinetochore interactions by phosphorylating microtubule-binding components within the kinetochore.
Chromosomal Passenger Complex
monitors microtubule attachment at kinetochores
The first ensures that kinetochore-microtubule attachments are weak until bi-orientation occurs, ie biorientation stabilizes kinetochore-MT attachment.
includes Ndc80 and Aurora B kinase
The second promotes tight microtubule-kinetochore association after biorientation
includes protein phosphatase 1
Aurora B Kinase
Phosphorylates Ndc80 preventing tight association with a microtubule.
Protein Phosphatase 1 (PP1)
An outer kinetochore protein, dephosphorylates Ndc80. This allows strong microtubule-kineotchore associations.
Aneuploidy
Any deviation from the normal diploid number of chromosomes in which extra copies of one or more chromosomes are present or one of the normal copies is missing.
If anaphase occurs before all chromosomes are bioriented an incorrect number of chromosomes will segregate to daughter cells.
Anaphase A
Chromosome movement toward poles is powered by microtubule shortening and sustained attachement of the microtubule tips to kinetochores
Anaphase B
Movement of poles and lengthening of cell
Model organisms for cell cycle study
budding yeast (Hartwell)
fission yeast (Nurse)
sea urchins (Hunt)
frog embryos
Maturation Promoting Factor
Also known as mitosis promoting factor
Discovered from Masui and Market injecting the cytoplasm of recently fertilized frog eggs into undertilized eggs. This induced them to enter
Cyclin Dependent Kinases (CDKs)
phosphorylate and regulate activity of stage specific proteins
positive and negative feedback first enhances CDK activity and eventually promotes inactivation leading to abrupt transitions in cell cycle stage.
Cyclins
regulatory protein that bind and activate CDKs during the appropriate stage of the cell cycle and determine their substrate specificity.
Regulation of cell cycle transitions
A) activity of stage specific CDKs at different points in the cell cycle- though all CDKs are always present, they are only active when bound by partner cyclins- this panel also shows the activity of a key regulator of entry into anaphase and exit from the cell cycle called APC
B) the expression of the various cyclins at different points in the cell cycle
C) the kinase activity of the various cyclin/CDK complexes through the cell cycle
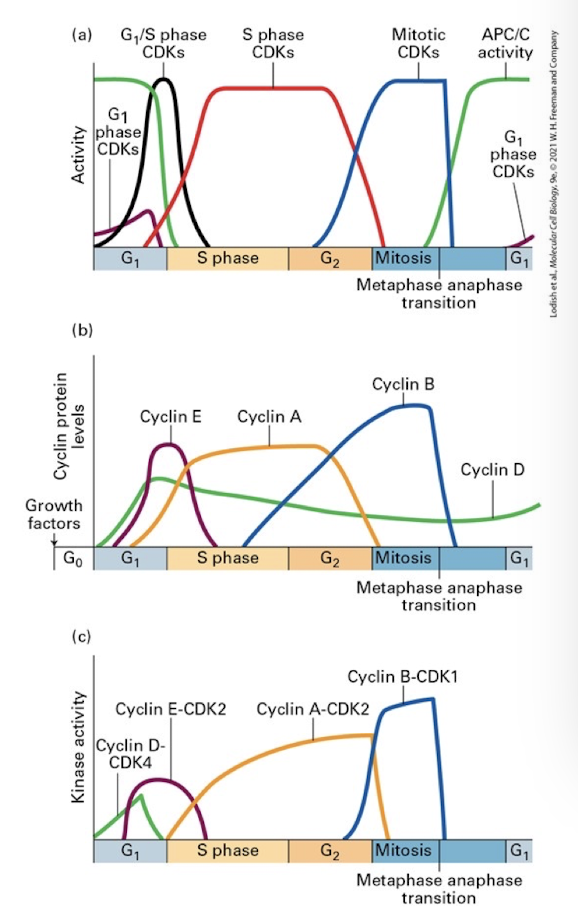
Anaphase Promoting Complex (APC)
A ubiquitin ligase that targets securing, mitotic cyclins, and other proteins for proteasomal degradation from the onset of anaphase until entry into the subsequent cell cycle.
Not a cyclin-CDK complex, but is a ubiquitin ligase complex that triggers exit from mitosis (as well as promotes the metaphase to anaphase transition)
Promotes the degradation of:
A) Anaphase inhibitory proteins to promote separation of sister chromatids at the anaphase to metaphase transition
B) As well as S phase and mitotic cyclins to promote exit from M phase and reentry into Interphase.
SCF
Ubiquitin-protein ligase that ubiquitinylates inhibitors of S-phase CDKs and many other proteins, marking them for degradation by proteasomes.
Functions to degrade inhibitors of S phase promoting proteins. It therefore helps trigger the G1-S phase transition
Active SCF promotes degradation of G1 and G1/S phase cyclins and CDK inhibitory proteins
CDK activating kinase (CAK)
After being bound by a cyclin, each CDK is activated via phosphorylation by CAK
Phosphorylates CDKs on a threonine residue near the active site. This phosphorylation is essential for CDK activity.
Inhibitory Phosphorylation
Most CDKs also have residues that are phosphorylated by inhibitory kinases. This pauses the CDK after CAK phosphorylation- allowing rapid activation at the appropriate time via the action of phosphatase.
CDK Inhibitors (CKIs)
Prevent premature activation of S and M phase CDKs
These are allosteric regulators prevent cells with damaged DA from passing the restriction point, and in doing serve as major roadblocks to tumor formation.
Mitogen
Any extracellular molecule, such as a growth factor, that promotes cell proliferation.
E2F
A TF family that will promote entry into S phase
G1 cyclin-CDKs indirectly activate this. During G1 E2Fs are bound to their target genes but are inhibited from promoting transcription by a negative cell cycle regulator Retinoblastoma (Rb)
Retinoblastoma (Rb)
negative cell cycle regulator
a transcriptional repressor that must be removed in order for the cell to progress into S phase.
recruits deacetylases and methyltransferases, which make the DNA inaccessible to transcription by promoting heterochromatin.
Rb is inactive in many cancer cells (either by mutation or hyperphosphorylation due to mutation in other genes.) This allows proliferation even in the absence of growth factors.
G1/S phase GDKs prepare DNA for replication
Turning off APC- which otherwise promotes S phase cyclin degradation
The two functions, and stage specific targets, of this ubiquitin ligase are mediated by distinct allosteric activators
During late anaphase the protein Cdh1 binds APC giving it specificity for S and M phase cyclins- this promotes exit from mitosis and reentry into interphase
When G1/S phase cyclins accumulate and activate their CDKs, they phosphorylate Cdh1 which dissociate from APC
APC can no longer target S phase cyclins for degradation, and they rise to threshold levels to promote S phase entry.
Inducing degradation of a CKI that inhibits S phase CDKs
As S phase cyclin-CDks accumulate they are bound and inactivated by a CKI this CKI was first identified in yeast and named Sic1.
When G1/S-cyclins reach a threshold level they phosphorylate Sic1 (targeting it for poly-ubiquitination by SCF). S-phase cyclin-CDKs can now activate proteins necessary for DNA replication
Degradation of an S phase CDK Inhibitor Triggers DNA Replication
The mammalian homolog of Sic1 named p27 which inhibits bothe S-phase and G1/S CDKs
Two mechanisms of p27 inactivation
Like Sic1, p27 is also phosphorylated by high G1/S and S phase CDKs
MAPK- activated via RTK transduction by growth factors- phosphorylates and activates p27
Both events promote poly-ubiquitination proteosomal degradation of p27
Origin of recognition complexes
Initiates DNA replication that occurs throughout S phase
No ORC initiates replication more than once
MCM helicase
During early G1, when S phase cyclin-CDK levels are low, ORC recruits MCM helicase to form pre-replication complex, but remains inactive.
Once S phase cyclin-CDK reach their peak, they phosphorylate ORC cofactor proteins and MCM helicase
this releases ORC and recruits several MCM helicase activators as well as the DNA polymerases
Entry in Mitosis
When S phase is complete cells must:
Radiate the mitotic spindle
Breakdown the nuclear envelope
Restructure or modify almost all organelles
Capture chromosomes on kinetochore microtubules
Segregate sister chromatids to opposite poles of the spindle
All of these events are triggered and controlled by the mitotic cyclin-CDK complexes.
CDK-activating Kinase (CAK)
Phosphorylates CDKs on a threonine residue near the active site. This phosphorylation is essential for CDK activity
Wee1
protein-tyrosine kinase; phosphorylates CDKs on threonine 14 and tyrosine 15 to inhibit CDK activity
an inhibitory kinase which also phosphorylates CDK1 on conserved tyrosine and threonine residues- this pauses the complex and prevents it from phosphorylating target proteins until those phosphates are removed
Is a dual-specificity kinase (it can phosphorylate both tyrosines as well as serine/threonines).
Promotes early division by skipping G2, therefore smaller cells.
Cdc 14 phosphatase
A dual-specificity protein phosphates that triggers mitotic CDK inactivation at the end of mitosis.
Cdc25B phosphatase
One of a pair of dual-specificity phosphatases, along with Cdc25C, that dephosphorylates CDKs on threonine 14 and tyrosine 15, thereby activating CDKs
Cdc25C phosphatase
One of a pair of dual-specificity phosphatases, along with Cdc25B, that dephosphorylates CDKs on threonine 14 and tyrosine 15, thereby activating CDKs.
Cdc25 (aka String)
dual specificity phosphatase which removes the inhibitory phosphorylation on CDK1.
positive and negative feedback regulation:
Active mitotic CDK1 phosphorylates and stimulates Cdc25, further promoting its own activation
Additionally, CDK1 phosphorylates Wee1 leading to its ubiquitin mediated degradation by the SCF complex
Prevents division, but allows the cell to continue growing in an extended G2 phase. They are essentially trapped in G2 and therefore grow into long (string-like) cells.
Lamins
A group of intermediate filament proteins that form a fibrous network, the nuclear lamina, on the inner surface of the nuclear envelope.
Nucleoporin
Large group of protein that make up the nuclear pore complex. One class (FG-nucleoporins) participates in nuclear import and export.
Mitotic CDKs Promote Nuclear Envelope Breakdown
Active mitotic CDKs phosphorylate specific serine residues in all three nuclear lamins causing depolymerization of lamin intermediate filaments and disintegration of the lamina promoting disassembly of the nuclear envelope .
Mitotic CDKs also phosphorylate specific nucleoporins, causing nuclear pore complexes to dissociate into subcomplexes during prophase.
Phosphorylation of integral membrane proteins of the inner nuclear membrane is thought to decrease their affinity for chromatin and further contributes to the disassembly of the nuclear envelope and its collapse into the ER.
Aurora Kinases
Serine/Threonine kinases that play a crucial role in cell division by controlling chromatid segragation. Aurora B kinase destabilizes faulty microtubule-kinetochore interactions by phosphorylating microtubule-binding components within the kinetochore.
Compaction
Condensation of chromosomes and untangling of sister chromatids (aka sister chromatid resolution) Reduces chromosome length up to 10,000x
without compaction DNA would become entangled and break as the chromosomes segregate during anaphase
Condensins
Protein complex that promotes chromosome condensation
loops each individual chromatid arm into the dense, recognizable M-phase chromosomes, still joined at the centromeres by cohesin.
Topoisomerase II
As condensins work to compact the chromosomes entanglements (knots) between sister chromatids are resolved by topoisomerase II enzymes
Cleaves one sister chromatid, passes the other sister chromatid through this break and then re-ligates the cut ends back together