Key Points About Methods Paper #3
1/35
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
36 Terms
cGAS
Cyclic GMP-AMP synthase
CBASS
Cyclic-olgionucleotide-based anti-phage signaling system
CD-NTase
cGAS/DncV-like Nucleotidyltransferase, a large family of enzymes found in animals and bacteria.
These enzymes play a crucial role in cellular signaling, in response to pathogens and changing environmental conditions.
Ubiquitination cascade (humans)
Activating enzyme E1
Conjugating enzyme E2
Ligating enzyme E3
What can the Ubiquitin-Ligating Enzyme do?
Ubiquitin-Ligating Enzyme E3 can covalently link ubiquitin to a substrate.
The deubiquitinating enzyme can remove ubiquitin from a substrate.
What is special about Type II CBASS?
Has no E3 ligase
E1/E2/JAB-CBASS
What is the most important point about E2-CBASS that was discovered in the paper?
E2-CBASS uses a more concise and specific mechanism, with a distinct protein chemistry, to regulate anti-phase signaling, compared to E1E2/JAB-CBASS.
What is cGAMP?
A cyclic guanosine monophosphate-adenosisne monophosphate a signaling molecule involved in the innate immune response of certain organism.
It is a second messenger, receives signal from the surface of the cell to target molecules within the cell.
Anisotropic
A material or object that exhibits properties that differ depending on the direction of measurement
Abortive Infection
The bacterial cell self-destructing
Abortive infection prevents the virus from multiplying and spreading to other bacteria
2 main proteins in the CBASS system
cGAS – makes the alarm signal molecule
E2 – activates cGAS
How can E2 activate cGAS?
By linking multiple proteins together into a chain via poly-cGASylation
Steps E2 takes to link multiple cGAS proteins together
Processing – E2 acts like molecular scissors, snipping off the last two amino acids from the tail of a cGAS protein.
Conjugation – E2 forms a temporary covalent bond (thioester) with the newly exposed tail of the cGAS protein
Ligation – E2 transfers the cGAS it’s holding to another cGAS protein forming a strong permanent link (isopeptide bond) which creates the cGAS chain.
What is produced after the formation of the cGAS chain?
Once the chain is made the chain produces large amounts of the alarm molecule cGAMP.
The cGAMP signal activates an effector protein. The effector protein goes on to disrupt the bacterial cell’s membrane causing the cell to die and stop the phage infection.
Purpose of identification tags?
To make purification and detection of the specific proteins the plasmids produced easier
What are two examples of tags used during the detection?
His-tag (binds to nickel) and Maltose Binding Protein (MBP) tag.
What chemical was used to make the plasmids put into e.coli start production the target proteins?
IPTG
How was bonding studied?
cGAS and E2 were purified together to see how they form a covalent complex over time.
How was cleavage tested?
They formed cGAS - E2 complex in a low ph (acidic) condition to see if the covalent bond would break.
How were the results for the bonding and cleavage experiments were visualized?
SDS-PAGE, a technique that separates proteins by size on gel
The setup of Mass Spec?
The protein complexes were cut into smaller pieces called peptides and then measured to find the exact mass of each piece.
What was learned by comparing the exact masses of peptides from the cGAS-E2 complex to the individual proteins?
After they could pinpoint exactly which amino acids were forming the covalent bond
Where was the covalent bond located?
The link was between Y405 of cGAS and K159 of E2
Purpose of Edman Degradation
To confirm that E2 was cutting off the tail off cGAS. Edman degradation determines the sequence of amino acids at the beginning of a peptide (N)
Purpose of GREEN FLUORESCENT PROTEIN?
Used a GFP to see the chaining of cGAS leading to poly-cGASylation.
Each additional green band is one additional cGAS added to the chain.
Purpose of WESTERN BLOT?
To see if poly-cGAS was inside a bacterial cell.
A cGAS specific antibody was used to detect only the cGAS protein in the complex mixture.
Purpose of ALPHAFOLD AND COLABFOLD?
To generate a 3D model of what the cGAS-E2 complex looks like.
Purpose of Cryo-EM?
To get a high resolution image of the 3D structure of the molecules. CGAS – E2 was frozen in liquid ethane to trap the molecules in a thin layer of ice so that their natural shape was preserved.
Purpose of HPLC?
High performance liquid chromatography, to check the activity level of the complex. ATP and GTP were available to cGAS and HPLC measured how much cGAMP was produced.
Purpose PLAQUE ASSAY?
Proves if CBASS protects bacteria from phages. Bacteria was grown on agar and then drops of phage were added.
No defense = phages infect cells and creates plaques
Protection = CBASS works, triggers abortive infection, kills infected cells, no plaques c
Purpose of FLOURESCENT PROBES TO TEST PH
To see if pH was a sign of infection. After infection the pH of the cells dropped.
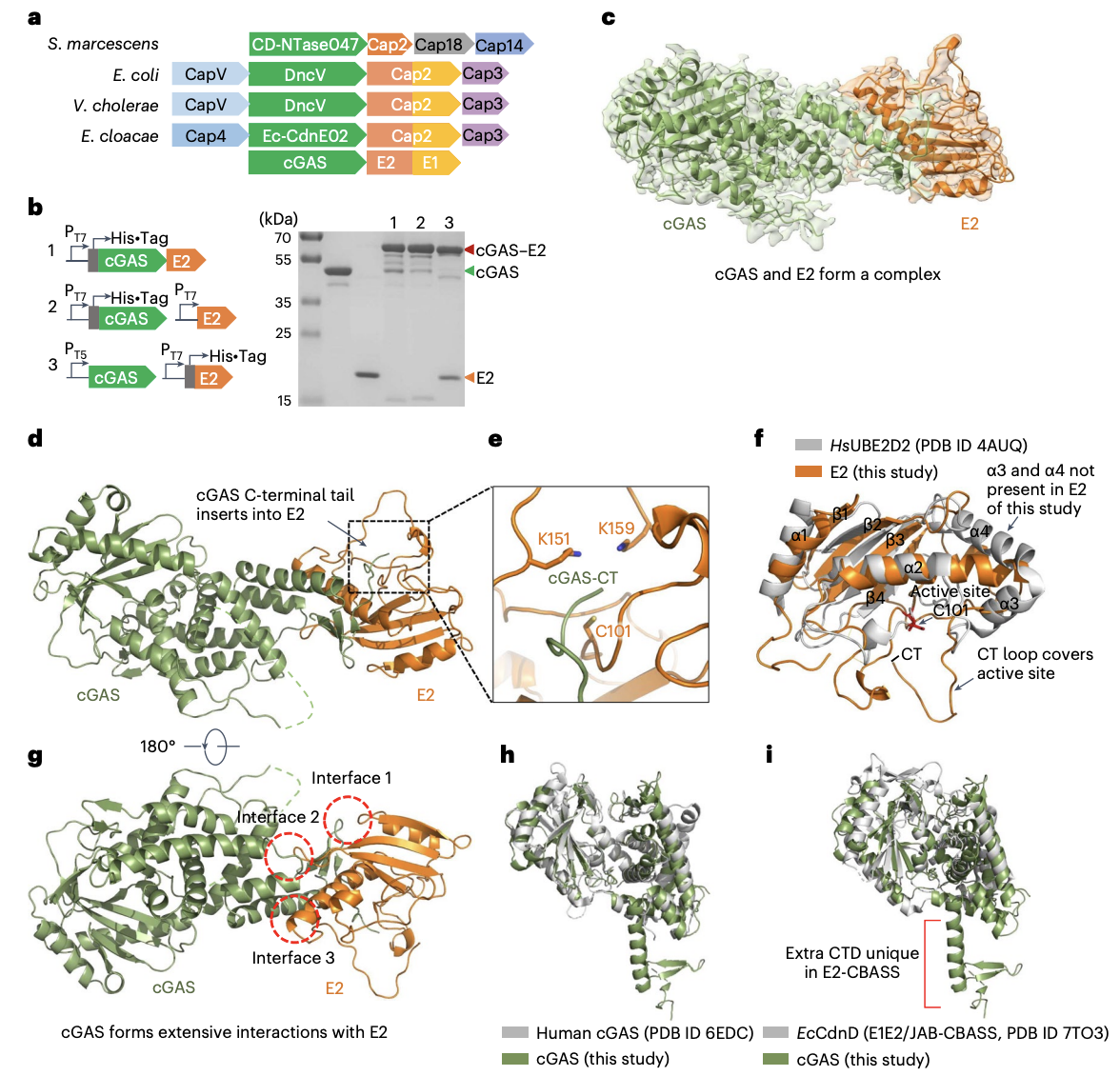
FIGURE 1: The cGAS and E2 Proteins Form a Unique Complex
A. Showing how different operons are between different species of bacteria. S. Marcescens is noticeably different because it contains genes for cGAS and E2.
B. Shows the SDS-PAGE gel. Shows the new cGAS-E2 complex making a new band. The bands size is the combined weight of cGAS and E2. Proves covalent bond since they didn’t separate under the harsh environment of the SDS-PAGE gel.
C. Shows Cryo-EM structure.
D.Detailed Cryo-EM. C-terminal tail of cGAS connects directly into E2 enzyme.
E. Zoomed in view to see location of covalent bond.
F – I: Compares E2-cGAS complex to similar structures in humans.
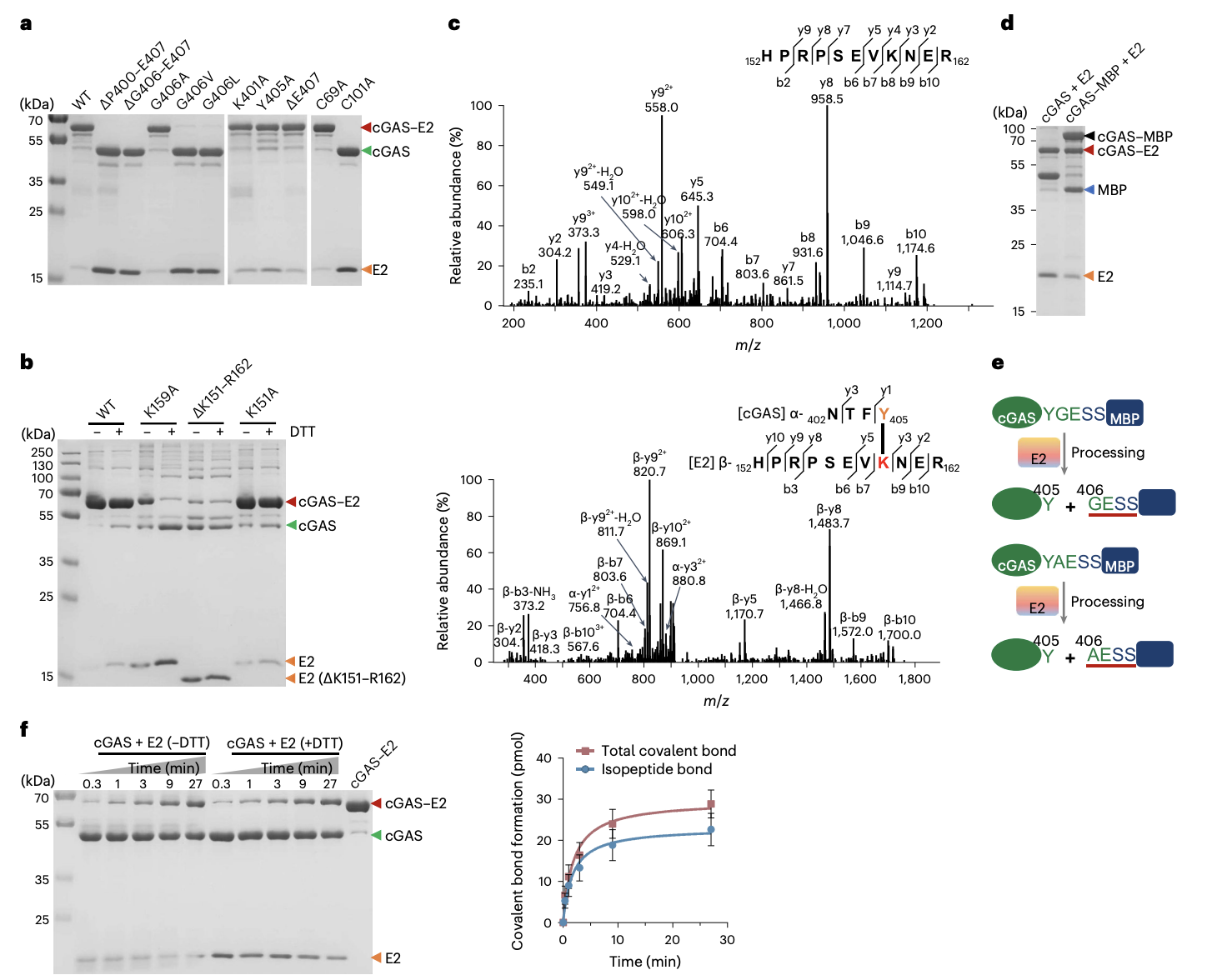
FIGURE 2: Identifying the Covalent Linkage Between E2 and cGAS
A. Shows an SDS-PAGE gel used to test which amino acids are essential for forming the cGAS-E2 complex. Several versions (mutants) of cGAS and E2 were created, each with a specific amino acid changed or deleted.
cGAS Mutants:
B. Deleting the last eight or the last two amino acids of cGAS prevented the complex from forming.
C. Changing the second-to-last amino acid, Glycine 406 (G406), to a bulky one like Valine or Leucine also broke the connection. This shows G406 is critical for recognition.
E2 Mutants:
D. Changing Cysteine 101 (C101) to Alanine completely abolished the bond. This identifies C101 as a crucial active-site residue.
The bond formation requires the C-terminal tail of cGAS (specifically G406) and a key cysteine (C101) in E2's active site
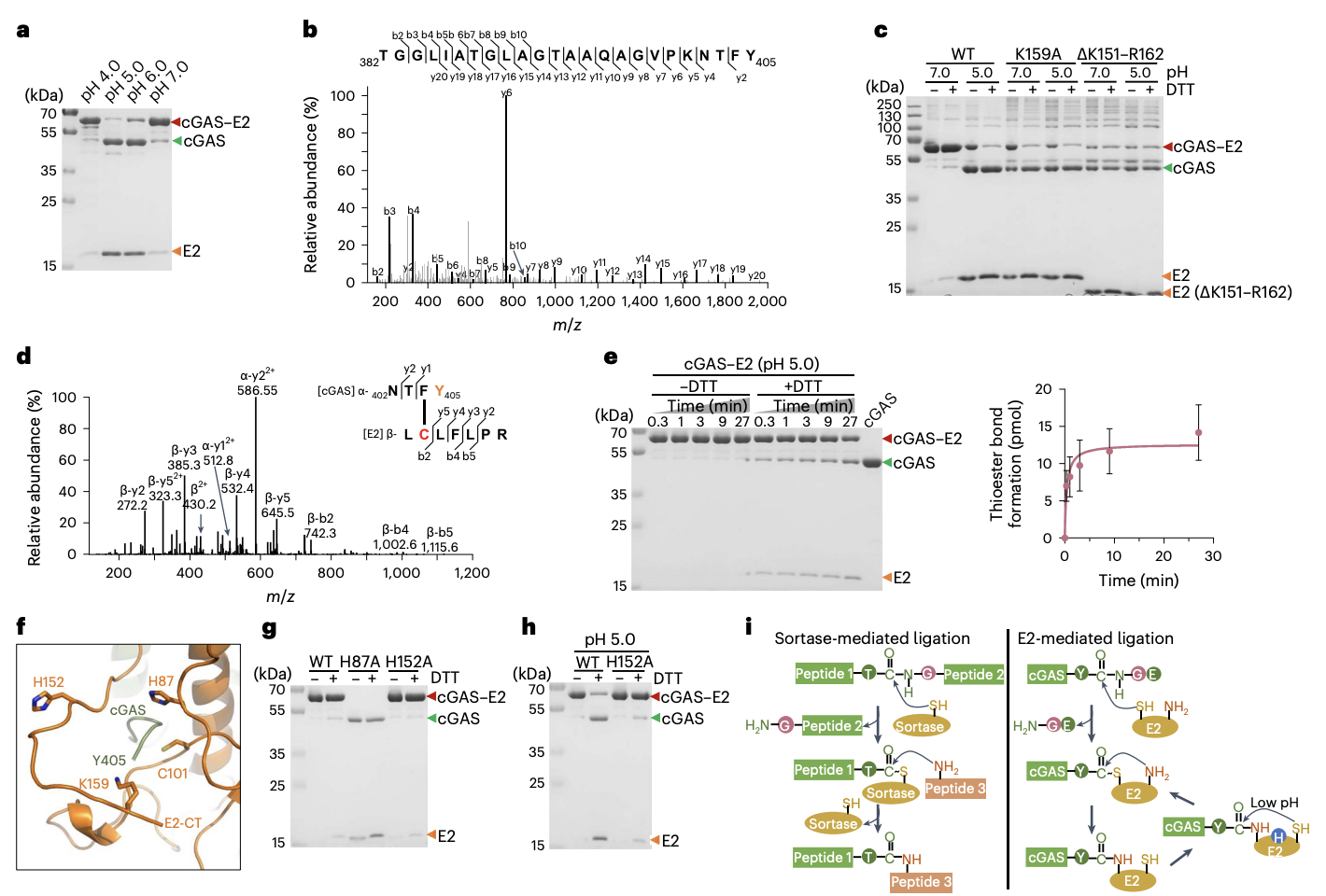
FIGURE 3: The function of E2 as a cysteine protease
DTT a chemical that specifically breaks unstable thioester bonds but not stable isopeptide bonds
Proof cGAS - E2 is held together by a DTT-resistant isopeptide bond
When a key Lysine residue on E2, K159, was mutated (K159A), the complex became much more sensitive to DTT
PANEL C: The analysis showed this "NTFY" fragment was attached specifically to the Lysine at position 159 (K159) of the E2 peptide.
This is direct molecular evidence that the final, stable link is an isopeptide bond between Y405 of cGAS and K159 of E2.
D/E: MBP tag is cleaved during reaction. Edman sequencing was used to identify the first amino aid released, the cut was revealed to be between Y405 and G406
The last few panels show how the proteins link over time, the cGAS-E2 complex takes around 30 minutes to form
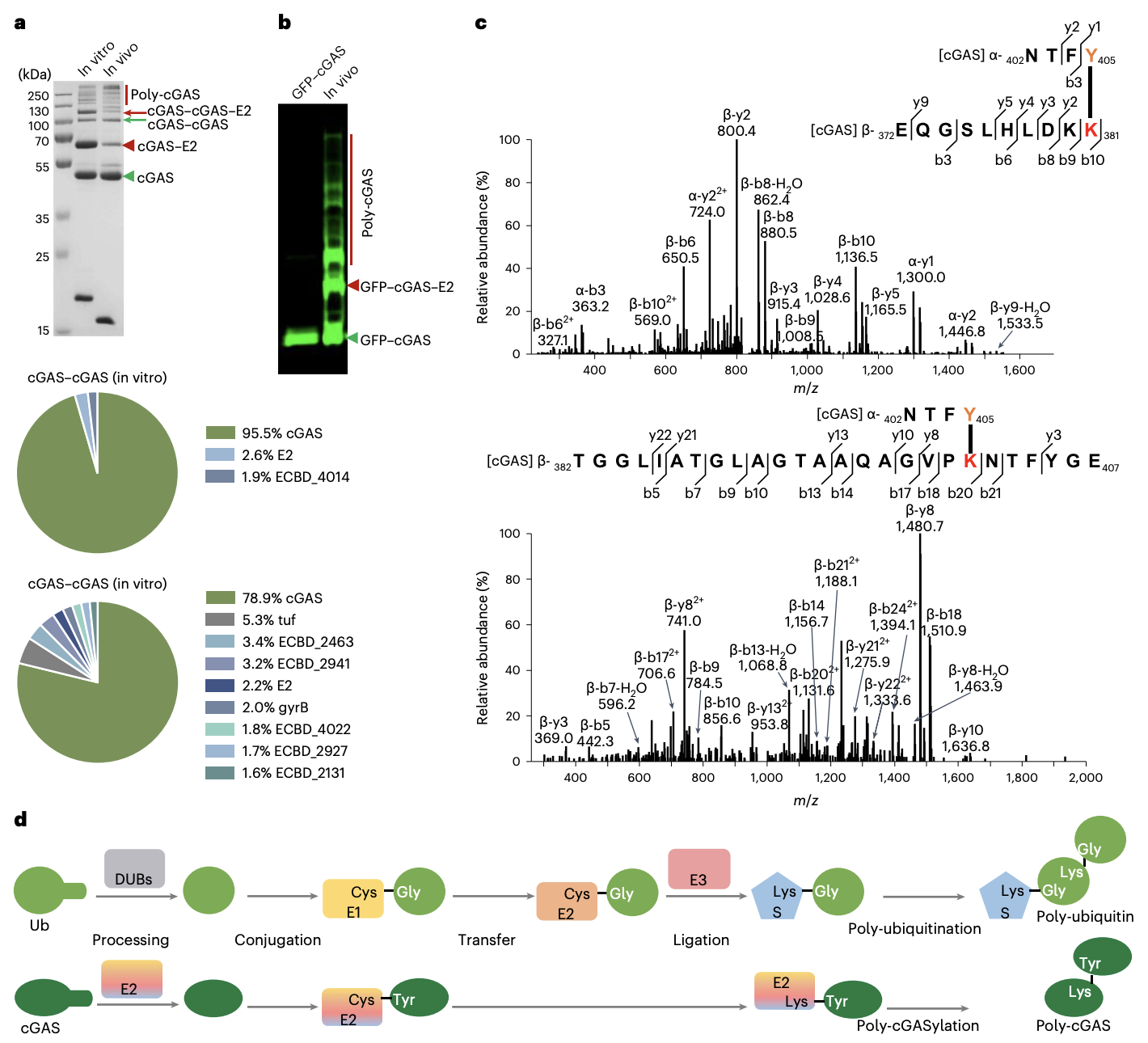
FIGURE 4: E2 Catalyses poly-cGASylation of cGAS
The formation of the cGAS chains, visualized by GFP.
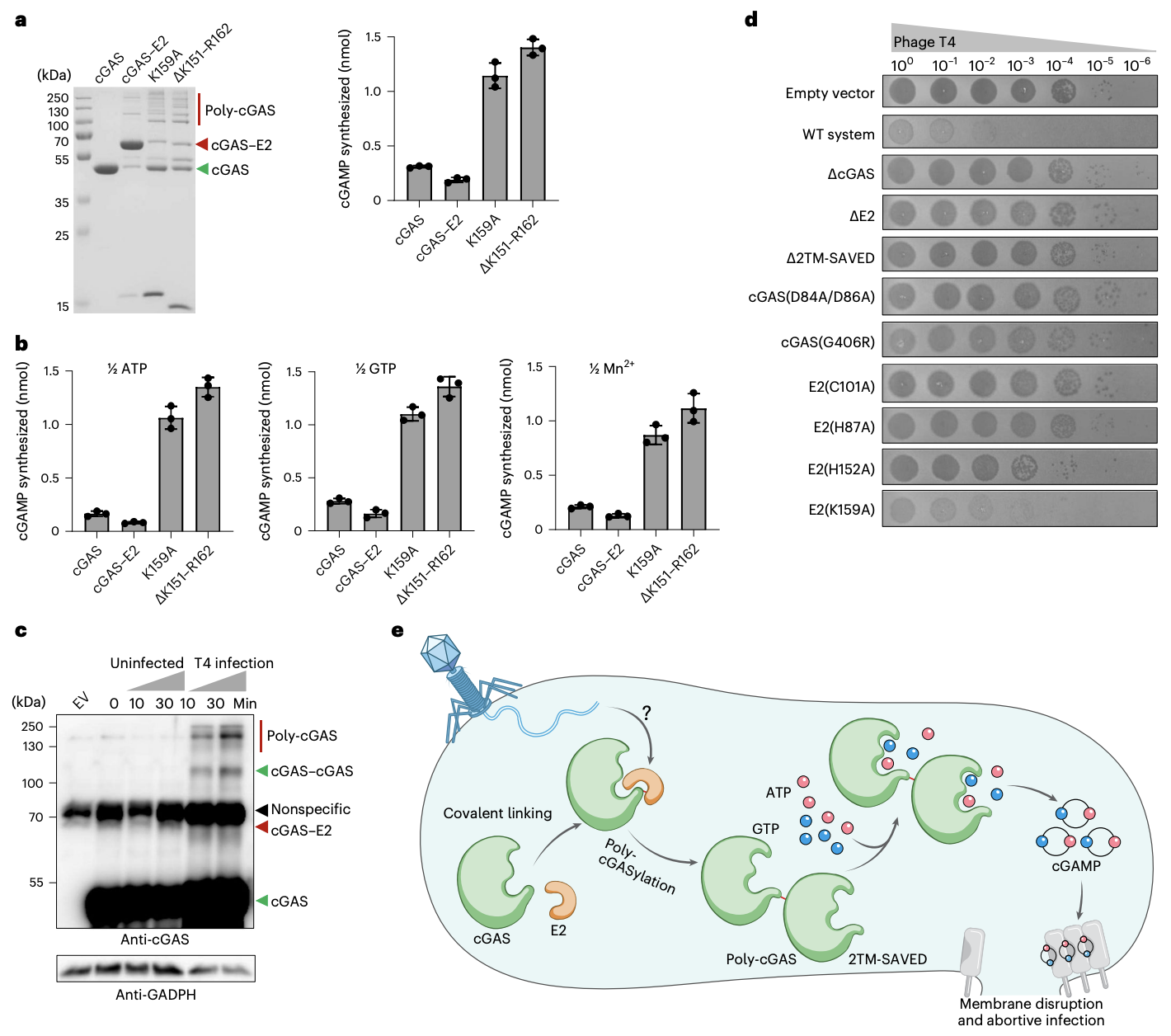
FIGURE 5: Poly-cGASylation stimulates cGAMP production and anti-phage immunity
An overview of the entire process. cGAS chain activation to cGAMP activation to abortive infection.