Microbial Metabolic Diversity: Catabolism
1/61
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
62 Terms
What are microbial tropisms for energy conservation?
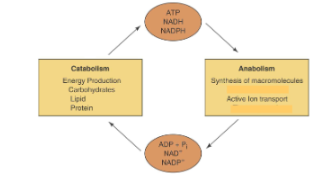
Metabolism is the sum of all chemical reactions in a cell, and it includes:
Catabolism: Energy-releasing reactions.
Anabolism: Energy-requiring reactions.
Microbial cells trap energy from the oxidation of inorganic compounds and/or light to ensure conservation of energy.
What is the reductionist approach in understanding metabolism?
The reductionist approach focuses on the following concepts:
Different organisms share similar chemical compositions and cellular structures.
They follow similar metabolic pathways to create, modify, and degrade components and structures.
Universal principles and pathways are key to understanding metabolism.
Universal energy carriers include ATP, NADH, NADPH, FADH2, and FMNH2.
When there is excess energy, organisms biosynthesise long-term energy storage molecules (in a more diverse range).
What is free energy in bioenergetics?
Free energy refers to the energy available to do work in a system. The change in free energy (ΔG) is used to determine the spontaneity of reactions.
What does ΔG represent in bioenergetics?
ΔG0′: Change in free energy under standard conditions.
ΔG: Actual free-energy change, which determines reaction direction
What do different values of ΔG indicate?
ΔG < 0: Exergonic, reaction proceeds spontaneously in the forward direction.
ΔG = 0: Equilibrium.
ΔG > 0: Endergonic, reaction proceeds spontaneously in the reverse direction.
How do catalysts affect reactions in bioenergetics?
Catalysts only affect the rate of a reaction, not the overall energy change (ΔG).
What is reduction potential (ΔE) and how is it linked to ΔG?
ΔE > 0: Redox half-reaction proceeds in the written direction (similar to ΔG < 0).
ΔE < 0: Redox half-reaction proceeds in the reverse direction.
The total ΔE is the sum of half-reactions.
How do microbial genomes contribute to redox reactions?
Microbial genomes encode many enzymes that allow a wider range of redox half-reactions, enabling more possible combinations for energy generation.
How do cells conserve energy from exergonic reactions?
Cells conserve energy by synthesising ‘high-energy’ or ‘energy-rich’ compounds that are biologically useful, using exergonic oxidation-reduction (redox) reactions to drive the synthesis.
What are the roles of electron donors and electron acceptors in redox reactions?
In redox reactions:
Electron donor: The substance that gets oxidized (loses electrons).
Electron acceptor: The substance that gets reduced (gains electrons).
What is reduction potential (E0′) in redox reactions?
Reduction potential (E0′) is the tendency of a substance to donate electrons. The more negative the E0′, the greater the tendency to donate electrons.
How is ΔE related to ΔG in redox reactions?
The ΔE of a redox reaction is related to ΔG via the Nernst equation, where a larger ΔE (more positive) results in more ΔG released.
How do substances with different E0′ values interact in redox reactions?
The reduced substance with a more negative E0′ donates electrons to the oxidised substance with a more positive E0′.
What happens when ΔE is large in a redox reaction?
The larger the ΔE (more positive), the more ΔG is released, indicating a more energetically favorable reaction.
How do electrons flow in metabolism?
In metabolism, electrons are not directly donated from reductants to oxidants. Instead, they are donated to coenzymes, which serve as intermediate ‘high-energy compounds’.
What coenzymes are used in catabolism and anabolism?
In catabolism: Coenzymes such as NAD/NADH, FAD/FADH2, and FMN/FMNH2 are used.
In anabolism: The coenzyme NADP/NADPH is used.
How do reduced coenzymes function in redox reactions?
Reduced coenzymes must donate their electrons to an electron acceptor so they can be reused in another redox reaction, undergoing cycling between their oxidised and reduced states.
What is the role of oxygen in the electron transport chain (ETC)?
In aerobic respiration, O2 serves as the terminal electron acceptor in the electron transport chain (ETC).
How do enzymes use cofactors in metabolism?
Many enzymes use cofactors to harvest electrons from a wide variety of inorganic and organic substrates, enabling a diversity of cellular redox reactions.
How is chemical energy stored in redox reactions?
Chemical energy released in redox reactions is primarily stored in certain phosphorylated compounds, including:
ATP (the prime energy currency)
Specific phosphorylated compounds, such as Phosphoenolpyruvate, 1,3-BPG, acetyl-P, ADP, etc.
How do Coenzyme A derivatives contribute to energy storage?
Hydrolysis of Coenzyme A derivatives, like acetyl-CoA, releases sufficient energy to form ATP from ADP and Pi.
How do reduced coenzymes contribute to energy transfer?
Reduced coenzymes help transfer energy by donating electrons in redox reactions, such as PEP + ADP → Pyruvate + ATP, which are favorable and used to synthesise ATP.
What is the role of ATP in metabolism?
ATP is used to phosphorylate compounds or is hydrolysed to drive endergonic reactions forward, playing a central role in energy transfer.
How is long-term energy stored in cells?
Long-term energy storage involves the biosynthesis of insoluble polymers that can later be oxidised to generate ATP.
What are examples of long-term energy storage in prokaryotes?
Examples include:
Glycogen (polyglucose)
Trehalose (diglucose)
Poly-β-hydroxybutyrate and other polyhydroxyalkanoates and lipids
Elemental sulfur (S)
Polyphosphate
Carbonates
Magnetic storage inclusions in magnetosomes
How do many microbes conserve energy?
Many microbes conserve energy via chemoorganotrophic growth, using the partial and full oxidation of organic compounds (catabolism).
What are the two major catabolic pathways in microbes?
Fermentation
Respiration
What is fermentation in microbial catabolism?
Fermentation is an anaerobic catabolic process in which an organic compound is partially oxidised by donating and accepting electrons, resulting in a fermentation end product.
What is respiration in microbial catabolism?
Respiration is a catabolic process in which an organic substrate is fully oxidised to CO2, with either O2 (aerobic) or another compound (anaerobic) as the terminal electron acceptor.
What is the common pathway for oxidation of glucose in microbes?
The almost universal pathway for glucose oxidation is Glycolysis (Emden-Meyerhof-Parnas or EMP pathway), which produces 2 Pyruvate, ATP, and reduced cofactors (such as NADH).
What alternative pathway do some microbes use for glucose oxidation?
Some microbes, like Pseudomonas aeruginosa, lack glycolysis and use the Entner-Doudoroff (ED) pathway to oxidise glucose, producing NADH and NADPH
What is the issue with redox balance in EMP and ED pathways?
Both EMP and ED pathways lead to a ‘redox imbalance’, where NAD(P)H must be re-oxidised to allow the degradation of another glucose molecule.
How is redox balance achieved in fermentative pathways?
In fermentative pathways, redox balance is achieved by one or more enzymatic reactions that oxidise NADH back to NAD, enabling continued glucose degradation.
Can fermentation pathways produce ATP?
Yes, some fermentation pathways allow for further ATP synthesis during the process.
How do ED-utilising microbes recycle NADPH?
ED-utilising microbes recycle NADPH to NADP in anabolic reactions.
What is the first step in microbial respiration after glucose oxidation?
After the partial oxidation of glucose to pyruvate via EMP/ED, the next step involves the formation of acetyl-CoA.
How is acetyl-CoA formed besides glucose oxidation?
Acetyl-CoA can also be formed from the oxidation of lipids and proteins.
Why is acetyl-CoA important in microbial respiration?
Many catabolic pathways converge at acetyl-CoA, just before its full oxidation in the TCA cycle.
What are the products of full oxidation of pyruvate in the TCA cycle?
Full oxidation of pyruvate in the TCA cycle leads to the biosynthesis of 3 high-energy compounds:
ATP/GTP
NADH
FADH2
Why can't microbes growing on C2 compounds like acetate use the TCA cycle?
they lack OAA regeneration options
Microbes growing on acetate avoid the full TCA cycle because it would result in excessive carbon loss as CO₂. Instead, they use the glyoxylate cycle, which bypasses decarboxylation steps and allows carbon conservation for biosynthesis.
What are the unique enzymes in the glyoxylate cycle?
The glyoxylate cycle includes two unique enzymes:
Isocitrate lyase
Malate synthase
How is the glyoxylate cycle similar to and different from the TCA cycle?
The glyoxylate cycle is highly similar to the TCA cycle, but it bypasses the decarboxylation steps, preventing the loss of 2 carbons in the cycle and allowing acetyl-to-acetyl additions, creating the C4 compound succinate.
What happens to succinate in the glyoxylate cycle?
Succinate produced in the glyoxylate cycle can be converted into OAA and subsequently used for gluconeogenesis (glucose synthesis).
Where does the glyoxylate cycle occur in eukaryotic microbes?
In eukaryotic microbes, the glyoxylate cycle occurs in organelles called glyoxysomes, a special type of peroxisome.
Why is inhibition of the glyoxylate enzymes important in therapy?
Inhibition of the unique glyoxylate enzymes offers new therapy options for treating multi-resistant microbial infections that rely on the glyoxylate cycle for growth in humans.
What happens during the full oxidation of glucose in microbial respiration?
The full oxidation of glucose (and other organic substrates) to CO₂ leads to the biosynthesis of many NADH and FADH₂ molecules
Why must cofactors like NADH and FADH₂ be re-oxidised?
Cofactors must be re-oxidised to allow the continued breakdown of glucose and other substrates in metabolism.
How does microbial respiration conserve energy?
Energy is conserved during re-oxidation of cofactors via respiration in the cytoplasmic membrane, using a multi-step transport of electrons in the electron transport chain (ETC)
What happens as electrons are transported in the ETC?
Protons (H⁺) are extruded to the outer surface of the cytoplasmic membrane, creating a proton motive force (pmf).
What factors contribute to the proton motive force (pmf)?
The pmf results from differences across the cytoplasmic membrane in:
Charge
pH
Electrochemical potential
What is the final stage of the electron transport chain (ETC)?
The final stage of the ETC, Complex IV, requires a terminal electron acceptor.
What is the terminal electron acceptor in aerobic respiration?
O₂ is the terminal electron acceptor in aerobic respiration, reducing to H₂O.
What happens under anoxic (oxygen-free) conditions?
Alternative electron acceptors are used in anaerobic respiration, such as:
NO₃⁻ (nitrate)
Fe³⁺ (ferric iron)
SO₄²⁻ (sulfate)
Fumarate
Why does anaerobic respiration conserve less energy than aerobic respiration?
None of the anaerobic electron acceptors have an E₀' as positive as the O₂/H₂O redox pair, meaning less energy is conserved.
What do different types of microbial energy conservation have in common?
Trophisms of energy conservation (aerobic & anaerobic respiration) converge in:
The electron transport chain (ETC)
The proton motive force (pmf)
Shared common principles of energy generation
What is ATP and how is it synthesised?
ATP is the ultimate high-energy compound in biological systems. It is synthesised via two main mechanisms:
Substrate-level phosphorylation – Direct transfer of a phosphate group to ADP from a high-energy substrate, occurring in glycolysis and the citric acid cycle.
Oxidative phosphorylation – ATP production using energy from the electron transport chain and chemiosmosis in the mitochondria, driven by a proton gradient.
What are the two main mechanisms of ATP synthesis?
Substrate-Level Phosphorylation (SLP) – Direct phosphate transfer from a high-energy compound to ADP.
Oxidative Phosphorylation – ATP synthesis via the Electron Transport Chain (ETC) and ATP synthase, powered by the proton motive force (pmf).
What is substrate-level phosphorylation (SLP)?
A process where high-energy phosphorylated compounds (e.g., phosphoenolpyruvate, 1,3-BPG) transfer their phosphate group to ADP, forming ATP
What is oxidative phosphorylation?
ATP synthesis driven by the ETC, where electrons flow through complexes, generating a proton gradient (pmf) that powers ATP synthase.
What is the role of ATP synthase?
ATP synthase is a membrane-bound enzyme complex that uses proton flow (pmf) to synthesise ATP
How does ATP synthase work?
F0 (membrane-bound motor): Protons flow through, creating torque (rotation).
F1 (catalytic unit): Torque drives conformational changes, capturing energy and producing ATP from ADP and Pi.
What powers ATP synthase?
The proton motive force (pmf), which results from protons being pumped across the membrane during ETC activity.