17: signalling through G protein-coupled receptors (GPCR)
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
25 Terms
what are GPCRs used for
to transmit signals that cant cross the plasma membrane
structure of GPCRs
7 transmembrane that spans across the lipid bilayer
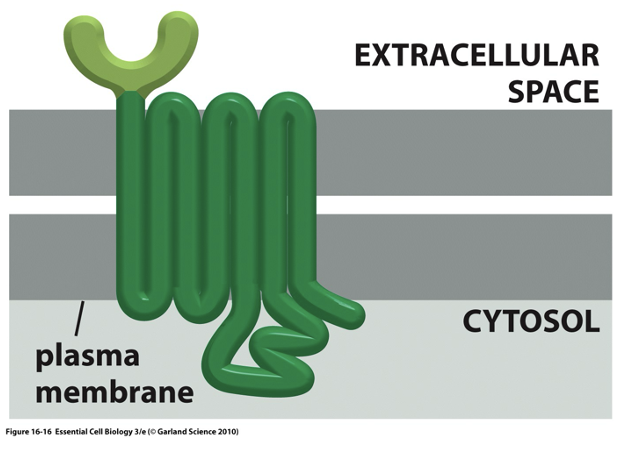
activation of GPCRs
activation by extracellular glands, eg:
glucagon
adrenaline
domaine
histamine
inactivation of GPCRs
inactivation by phosphorylation and internalisation
Signal transduction
process by which cells convert external signals, like hormones or neurotransmitters, into internal cellular responses
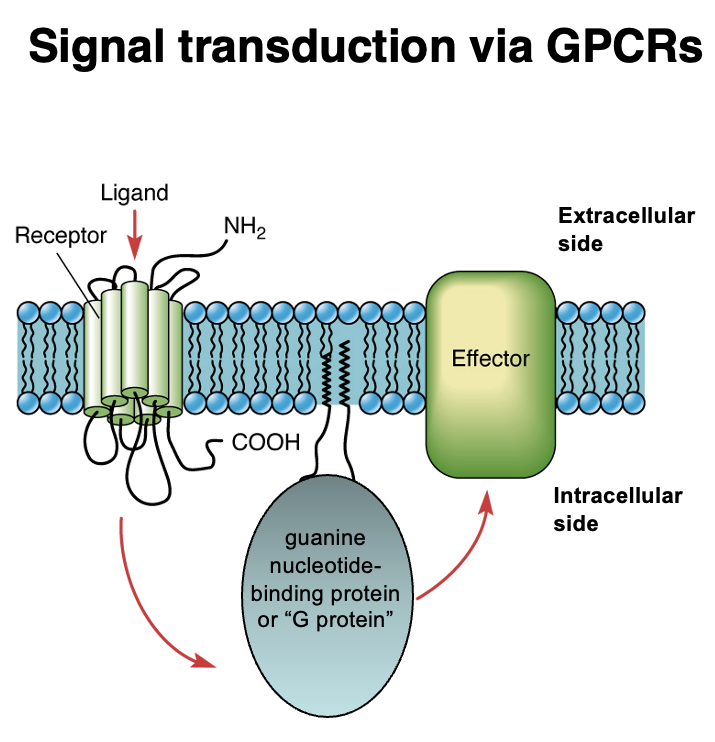
G-Protein Coupled Receptor Signaling
Steps in GPCR Activation:
1. Ligand Binding → Ligand binds to the extracellular domain of the GPCR.
2. GPCR Activation → Conformational change activates the G protein (guanine nucleotide-binding protein).
3. Effector Activation → The G protein interacts with an intracellular effector protein, triggering a signaling cascade.
4. Cellular Response → Signal transduction leads to physiological responses (e.g., gene expression, enzyme activation).
Key Components:
• GPCR (7-transmembrane receptor)
• G protein (GTP-binding protein, intracellular mediator)
• Effector protein (e.g., enzyme or ion channel)
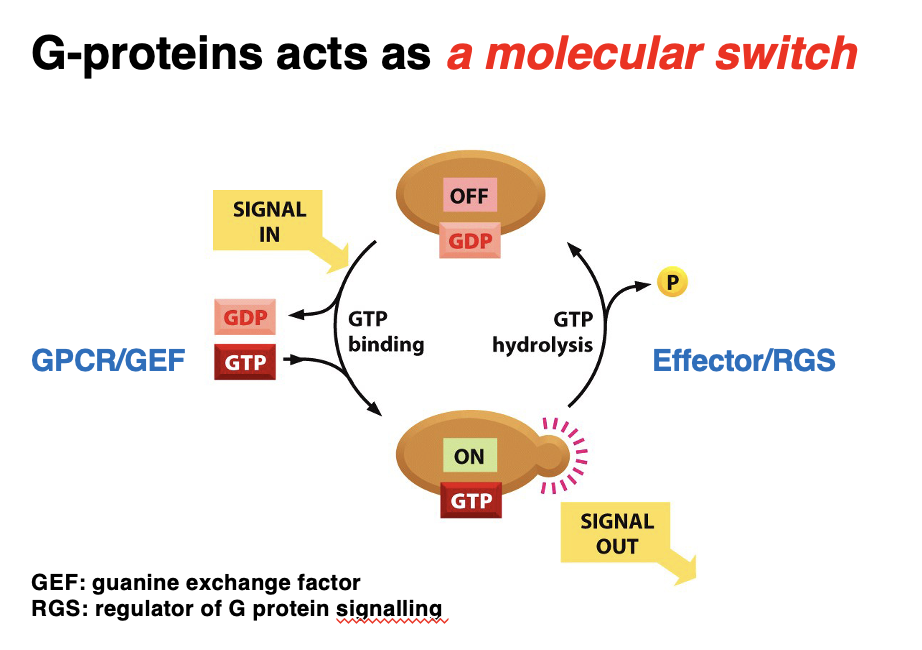
G-protein as a molecular switch
G-proteins toggles between active and inactive states in response to signals.
Activation Cycle:
1. OFF State (GDP-bound) → Inactive G-protein is bound to GDP.
2. Signal is received → GEF facilitates GDP-GTP exchange.
3. ON State (GTP-bound) → Active G-protein transmits the signal to effectors.
4. GTP Hydrolysis (via Effector/RGS) → RGS promotes GTP hydrolysis, turning G-protein off.
Key Regulators:
• GEF (Guanine Exchange Factor) → Activates G-protein by replacing GDP with GTP.
• RGS (Regulator of G-protein Signaling) → Inactivates G-protein by accelerating GTP hydrolysis.
G proteins: trimeric complex
In inactivated state, the G protein consists of a trimeric complex
3 subunits: alpha, beta, gamma
in activated state, the trimeric G protein complex dissociates
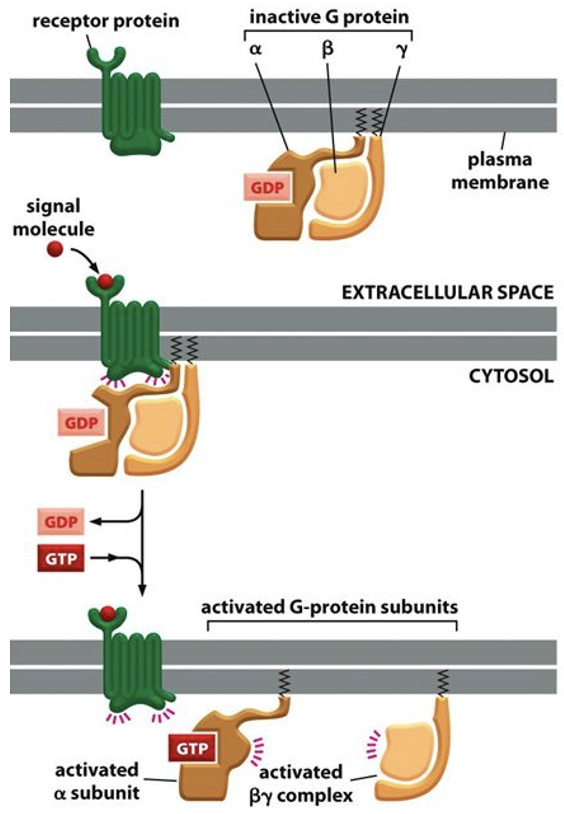
when trimeric G proteins ratite ion channels and membrane bound enzymes
ion channels: The interaction of G proteins with ion channels causes an immediate change in cell behaviour due to influx of ions entering/leaving cell
membrane bound enzymes: Enzyme activation via G proteins leads to production of intracellular signalling molecules - go on to act as second messengers
second messenger def
A rapidly produced diffusible signalling molecule that activates effector proteins.
diffusible = small molecule, can freely spread (diffuse) through the cytoplasm or within the membrane to reach and activate its target effector proteins.
In contrast, primary messengers (like hormones or neurotransmitters) usually stay outside the cell and signal via receptors.
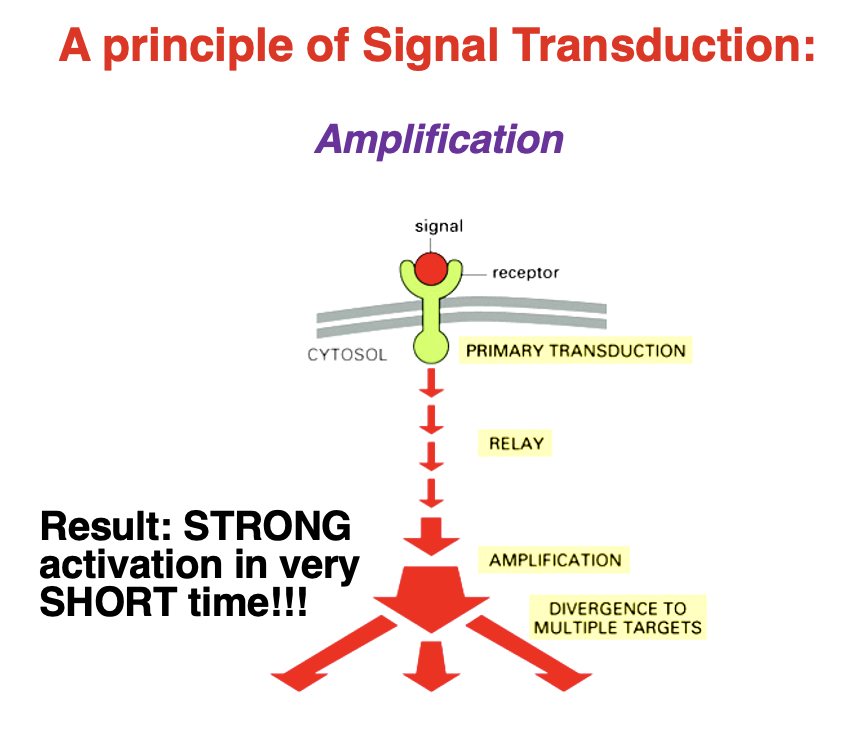
Signal Transduction: Amplification
• Amplification = allows a small initial signal to produce a large and rapid cellular response
Steps of Amplification:
1. Primary Transduction – Signal binds to a receptor.
2. Relay – Signal is passed to intracellular molecules.
3. Amplification – Enzymes or second messengers multiply the signal.
4. Divergence – Multiple pathways and targets are activated.
Result: A strong response in a very short time
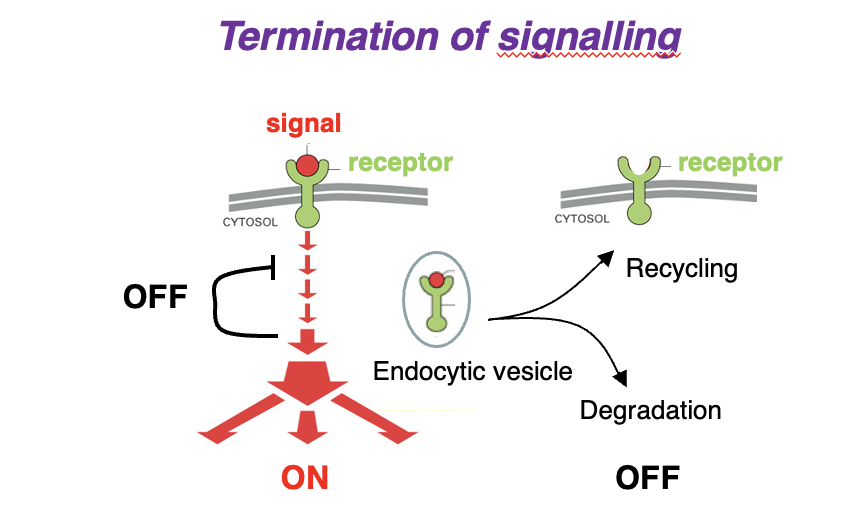
Signal Transduction: Termination of Signalling
1. Signal binds receptor → Activates signaling cascade (ON).
2. Signal & receptor internalized into an endocytic vesicle.
3. Two possible fates:
• Receptor Recycling: Receptor is returned to the membrane (OFF).
• Degradation: Receptor & signal are broken down (OFF).
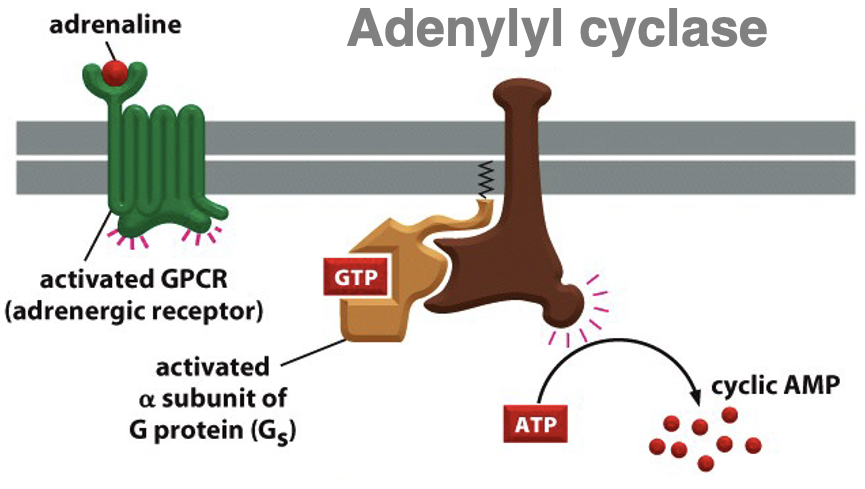
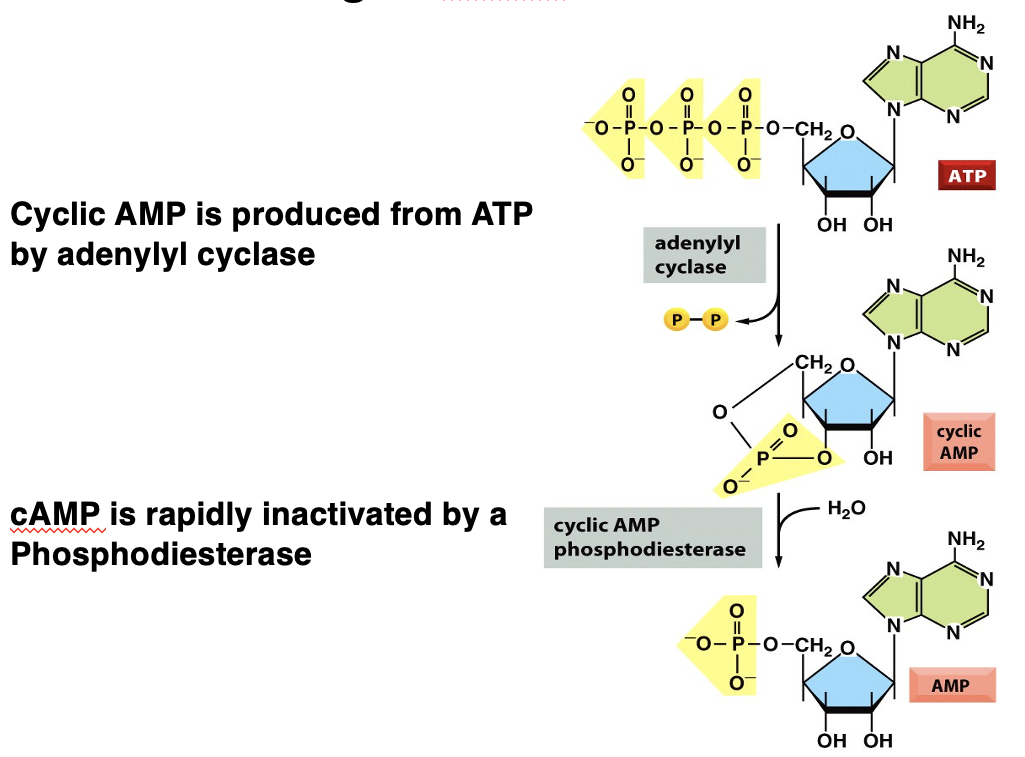
how is cAMP brought about
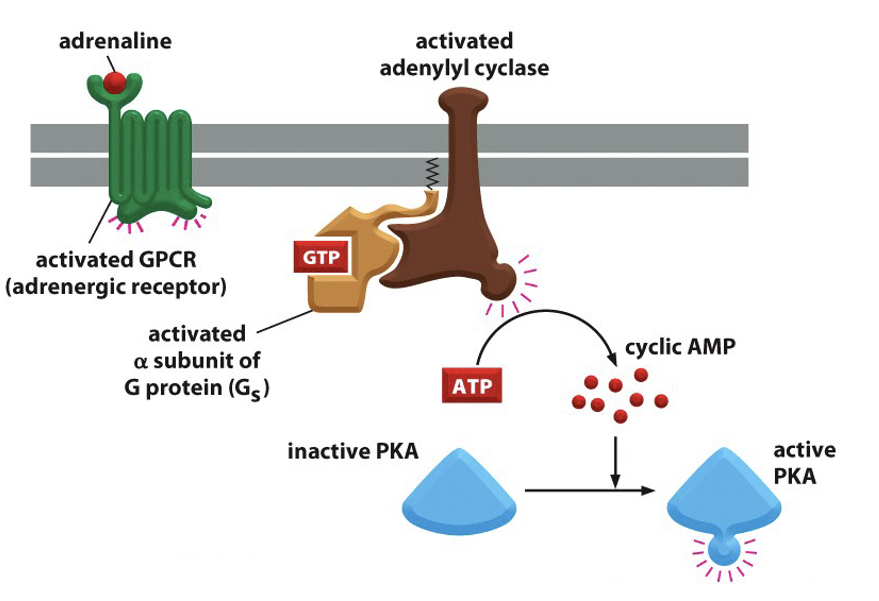
adrenaline binds to GPCR (adrenergic receptor) → activated GPCR
G protein activated by GDP replacement for GTP → subunits dissociate
alpha subunit binds to adenylyl cyclase enzyme → activated adenylyl cyclase
adenylyl cyclase converts ATP into cAMP (a secondary messenger)
inactivation of cAMP
phosphodiesterase hydrolyses cAMP, turning it into AMP
affect of cAMP on protein kinase A (PKA)
activates PKA
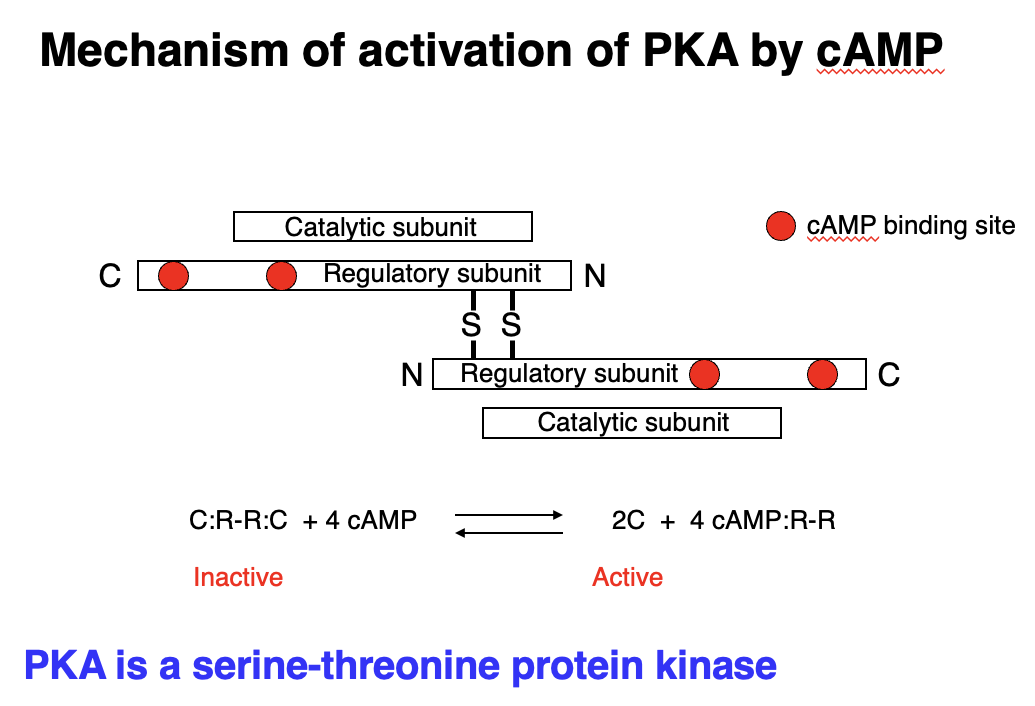
mechanism of activation of PKA by cAMP
• Protein Kinase A (PKA) = serine-threonine kinase - activated by cAMP.
• Inactive PKA: Exists as a tetramer (C:R-R:C), where regulatory subunits (R) inhibit catalytic subunits (C).
Activation Process:
1. cAMP binds to regulatory subunits.
2. Regulatory subunits change shape and release catalytic subunits.
3. Catalytic subunits (C) are now active, allowing them to phosphorylate target proteins.
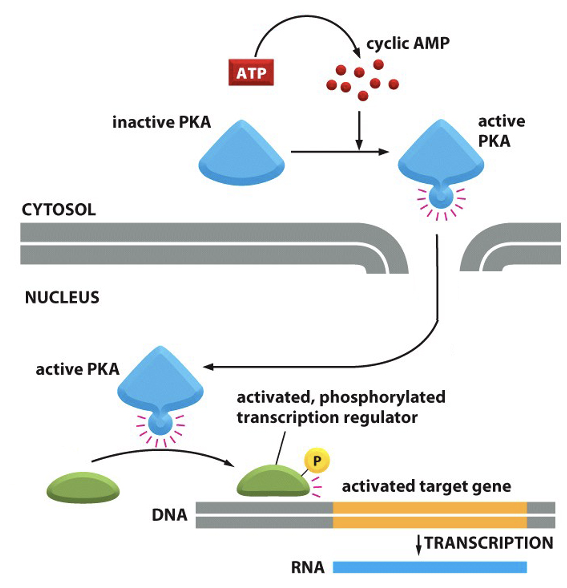
PKA and gene transcription
1. cAMP binds PKA, activating it.
2. PKA translocates into the nucleus.
3. PKA phosphorylates transcription factors (e.g., CREB).
4. Phosphorylated transcription factors regulate gene expression by increasing or suppressing transcription of target genes.
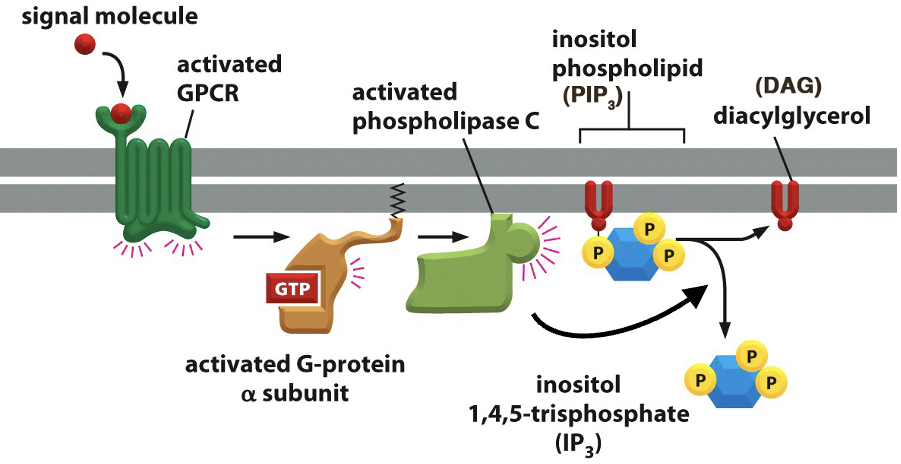
Second Messengers - DAG & IP₃
• Signal molecule activates GPCR, which in turn activates a G-protein (GTP-bound α-subunit).
• Activated G-protein stimulates phospholipase C (PLC).
• Phospholipase C cleaves PIP₂ into two second messengers: IP3 and DAG
• IP₃ (Inositol 1,4,5-trisphosphate): Triggers Ca²⁺ release from the ER.
• DAG (Diacylglycerol): Activates Protein Kinase C (PKC), leading to downstream signaling.
Inositol Phospholipids (PI)
key phospholipid involved in cell signalling.
Major Forms:
• PI = 80% of inositol-containing lipids
• PIP = Monophosphorylated form.
• PIP₂ = Precursor for second messengers IP₃ & DAG.
Function:
• Critical for signal transduction and membrane trafficking.
• Dynamically regulated by extracellular signals.
phospholipase C (PLC)
enzyme that cleaves phospholipids in the cell membrane, producing second messengers that regulate various cellular responses.
• PLC hydrolyses a specific membrane lipid called PIP₂
• generates two key signaling molecules:
1. IP₃ → Triggers Ca²⁺ release from the endoplasmic reticulum (ER).
2. DAG → Activates Protein Kinase C (PKC)
Types of PLC & Activation Mechanisms
1. PLC-β → Activated by GPCRs (G-protein-coupled receptors)
2. PLC-γ → Activated by Receptor Tyrosine Kinases (RTKs)
3. PLC-δ → Cytoplasmic form, regulated by calcium levels.
Functions independently but is enhanced by intracellular Ca²⁺.
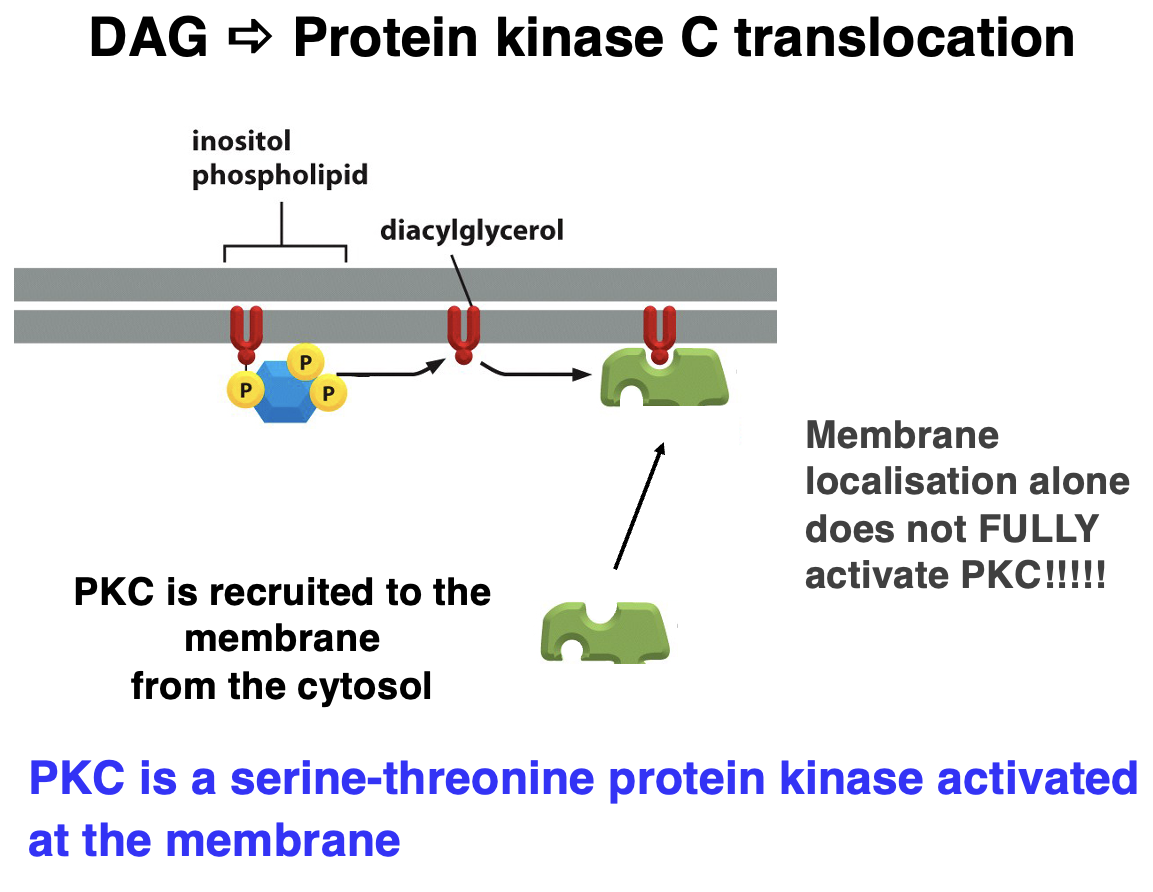
DAG ➠ Protein Kinase C (PKC) Translocation
• Diacylglycerol (DAG) = lipid-derived second messenger.
• activates PKC, but additional factors (like Ca²⁺) are required for full activation of PKC
Steps of PKC Activation:
1. Phospholipase C (PLC) Activation – Cleaves PIP₂ into DAG and IP₃.
2. DAG Stays in the Membrane – Acts as an anchor for PKC.
3. PKC Recruitment – PKC translocates from the cytosol to the membrane.
4. Full Activation – Requires Ca²⁺ and other cofactors.
5. Cellular Effects – PKC phosphorylates target proteins, regulating cell growth, metabolism, and gene expression.
PKC is a serine-threonine kinase activated at the membrane!
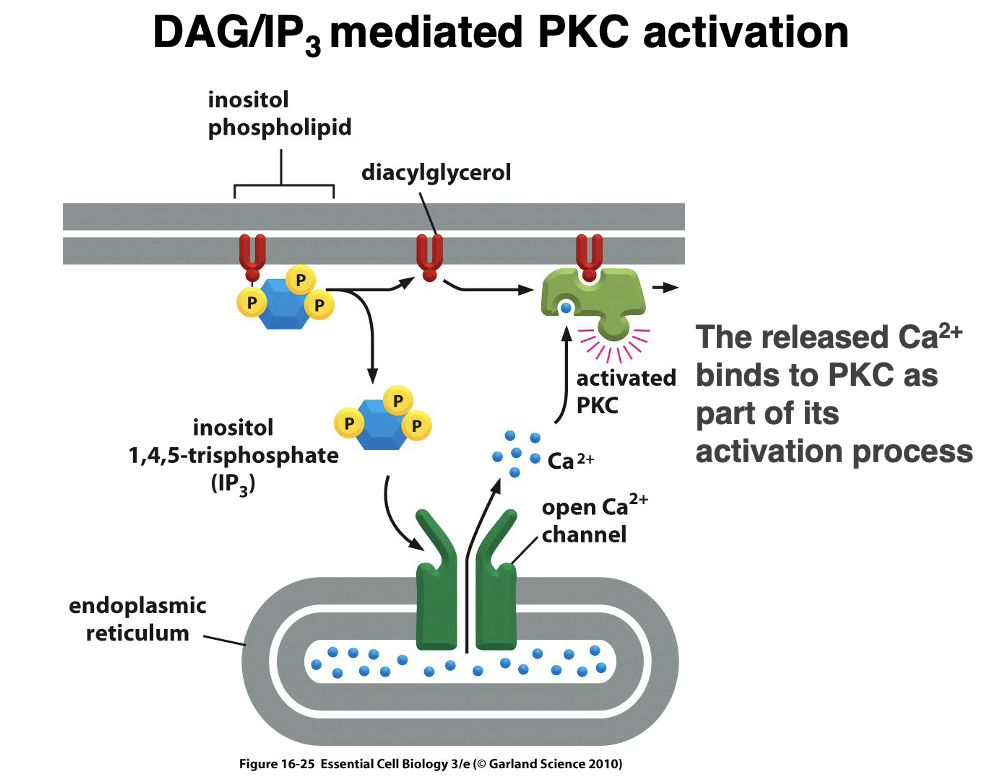
IP₃-Mediated Ca²⁺ Release
1. PIP₂ is cleaved by phospholipase C (PLC) into IP₃ + DAG (Diacylglycerol)
2. IP₃ diffuses through the cytoplasm and binds to IP₃ receptors on the endoplasmic reticulum (ER).
3. This triggers Ca²⁺ release from the ER into the cytoplasm, increasing intracellular calcium levels.
DAG/IP₃-Mediated PKC Activation
1. PIP₂ is cleaved by phospholipase C (PLC) into IP₃ + DAG (Diacylglycerol)
2. IP₃ binds to IP₃ receptors on the ER, leading to Ca²⁺ release into the cytoplasm.
3. Increased Ca²⁺ levels and DAG activate Protein Kinase C (PKC).
4. Activated PKC phosphorylates downstream targets, regulating key cellular functions such as gene expression, metabolism, and cell growth.
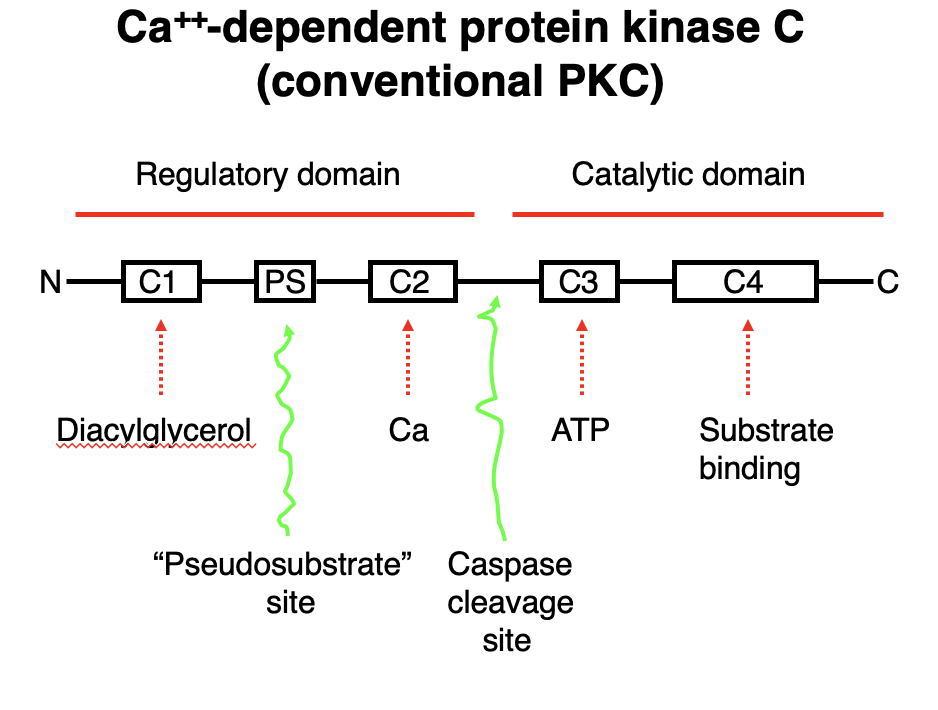
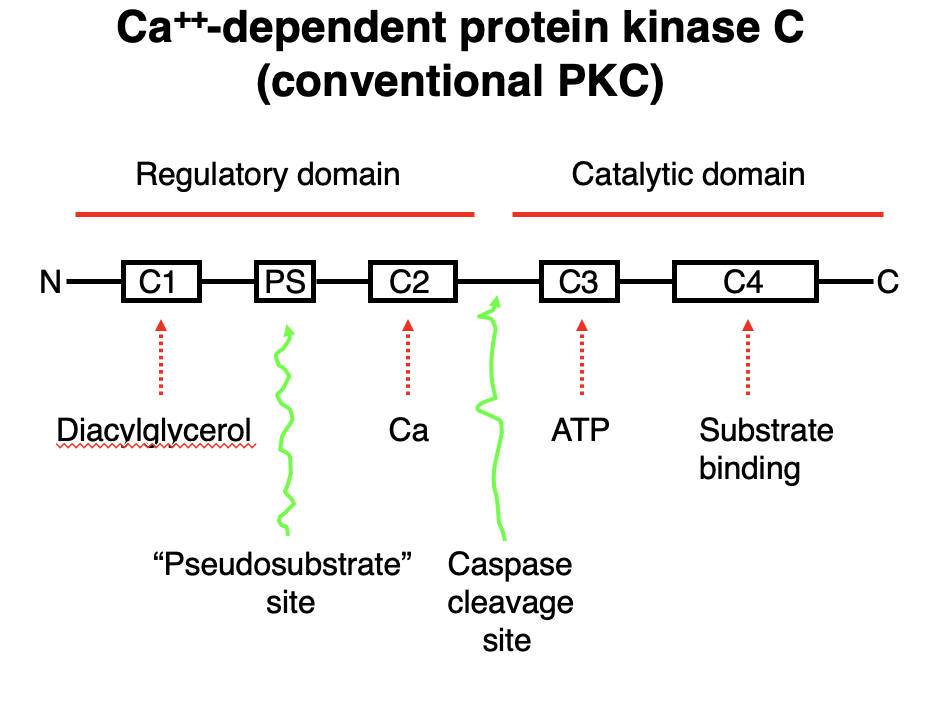
Ca²⁺-Dependent Protein Kinase C (Conventional PKC)
• Conventional PKC requires Ca²⁺, diacylglycerol (DAG), and ATP for activation.
Domain Structure & Function:
Regulatory Domain:
• C1 Domain → Binds Diacylglycerol (DAG).
• C2 Domain → Binds Ca²⁺, promoting membrane translocation.
• PS Site → Contains a pseudosubstrate region, keeping PKC inactive until activation.
Catalytic Domain:
• C3 Domain → ATP-binding site, necessary for phosphorylation.
• C4 Domain → Substrate-binding site, where PKC phosphorylates target proteins.
Activation Mechanism:
1. Ca²⁺ and DAG bind to PKC, relieving inhibition from the pseudosubstrate region.
2. PKC translocates to the membrane and undergoes conformational changes.
3. ATP binds to the C3 domain, allowing phosphorylation of substrates at the C4 domain.
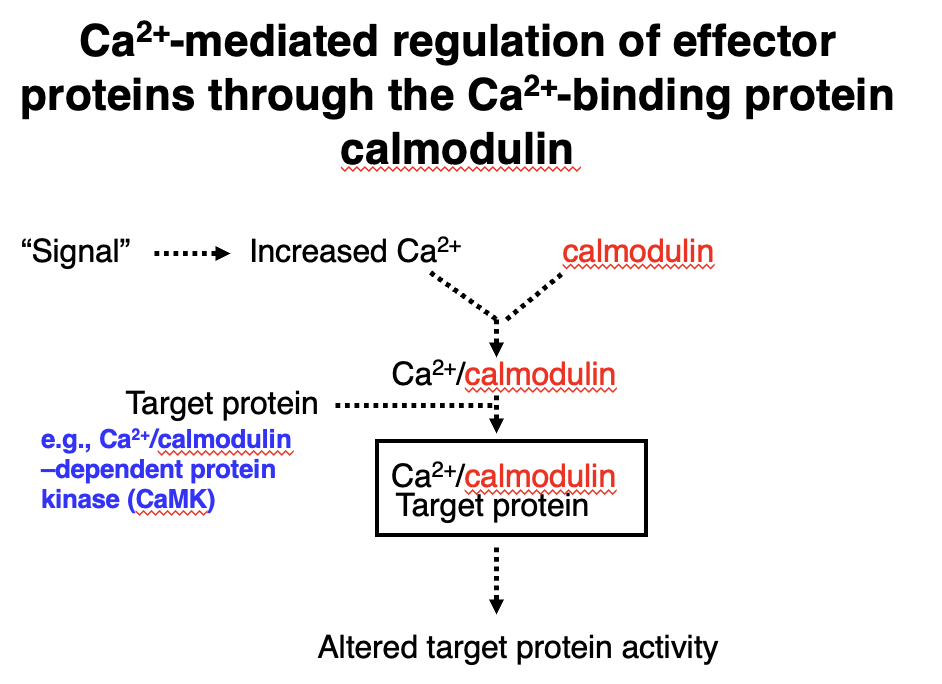
Ca²⁺-Mediated Regulation via Calmodulin
1. A signal triggers an increase in Ca²⁺ concentration.
2. Ca²⁺ binds to calmodulin, activating it.
3. Ca²⁺/calmodulin complex interacts with target proteins (e.g., Ca²⁺/calmodulin-dependent protein kinase, CaMK).
4. This interaction alters the target protein’s activity, leading to downstream effects.