IBI Exam 1
1/62
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
63 Terms
Ames test
developed in the 1970’s by Bruce Ames, Professor of Biochemistry at UC-Berkeley
fast, sensitive, easy, and inexpensive (it doesn’t use a live animal model) assay of the ability of a chemical compound or mixture to induce mutations in DNA
Measures the ability of a chemical to induce back mutations, or reversions, of an auxotroph to its original prototrophic state.
phototrophs
Bacteria that are able to synthesize all of the biochemicals needed for their own growth. They contain hundreds of metabolic pathways, each working to synthesize a single biochemical molecule.
The “wild type”, capable ofsynthesizing its own nutrients and growing on minimal medium
strain
a genetic variant or subtype of a microorganism
auxotroph
a particular strain or sample of bacteria unable to synthesize a needed molecule. In order for the bacteria to grow, the missing molecule must be added to the media.
The “mutant”, incapable of producing essential nutrients, and can only grow in complete media
reversions
“back mutations” of an auxotroph to its original phototrophic state
The greater the number of revertant colonies, the ___ mutagenic the test substance must be.
more
histidine
an amino acid needed for protein synthesis
limitations of the Ames test
the rate of mutagenicity isn’t the same in all organisms due to differences in the rate of chemical absorption by the cells and different metabolism of compounds
in some cases, the compound isn’t mutagenic but the metabolites of the compound are
compounds testing positive in this version of the Ames test would be subject to testing in mammalian model systems
metabolite
a substance formed during the metabolism of a compound
s. typhimurium strain TA1535
T to C missense mutation in the hisG gene, leading to a leucine to proline amino acid substitution
defect in the lipopolysaccharide cell wall causing them to be more vulnerable to exogenous mutagens
defective DNA excision-repair mechanisms, which would normally correct mutations arising during DNA replication or from exogenous mutagens
s. typhimurium strain TA1538
deletion of one base pair (c) in the hisD gene, causing a 1 frameshift mutation and bringing a stop codon into the reading frame prematurely
defect in the lipopolysaccharide cell wall causing them to be more vulnerable to exogenous mutagens
defective DNA excision-repair mechanisms, which would normally correct mutations arising during DNA replication or from exogenous mutagens
To quantitatively determine the mutagenicity of a compound, one must determine:
the number of mutant bacteria produced
the total number of cells exposed to the mutagen to begin with
binary fission
the process by which our bacteria doubled for about 10 hours
bacterial colony
consists of multiple microorganisms that are all from one mother cell which all gather together and are genetically identical
formula for CFU/mL
number of colonies * reciprocal of dilution factor / volume plated in mL
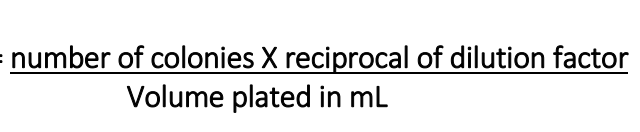
Serial dilutions: mL test sample, mL broth, dilution, dilution factor, reciprocal of dilution factor, amount plated (mL)
1.0 mL test sample, 9.0 mL broth
Dilution: 1:10, 1:100, 1:1,000, 1:10,000, 1:100,000, 1:1,000,000
Dilution factor: 10-1, 10-2, 10-3, 10-4, 10-5, 10-6,
Reciprocal of dilution factor: 101, 102, 103, 104, 105, 106
Amount plated: 1 mL
lawn
uncontrolled bacterial growth, which happens when we don’t reduce the concentration of a sample before plating it
culture media
Contains all the elements that most bacteria need for growth. Used for general cultivation and maintenance of bacteria.
Ex: Nutrient agar, LB broth, Trypticase soy agar
minimal media
A defined media that has just enough ingredients to support growth. Used to select for or against certain microorganisms
selective media
Contains or lacks a specific component that is needed to survive in that media. Used to separate specific microorganisms from others
ex: LB with ampicillin, HIS- media
differential or indicator media
Distinguishes one microorganism from another growing on the same medium. Uses biochemical characteristics microorganisms as indicators of their presence.
ex: Blood agar, Eosin Methylene blue, Mannitol salt agar
genotoxicity
the ability of a chemical to damage genetic information(which may lead to mutations and/or cancer)
If the reversion frequency of the tested product is at least 5 times higher than that of the Negative Control, then the tested product is genotoxic.
silent mutation
TTC → TTT
a point mutation that changes a codon but not the protein it produces
missense mutation
TTC → TCC
a point mutation that codes for a different protein
nonsense mutation
TTC → ATC
a point mutation that prematurely introduces a stop codon
insertion
a frameshift mutation in which a codon is added
deletion
a frameshift mutation in which a codon is removed
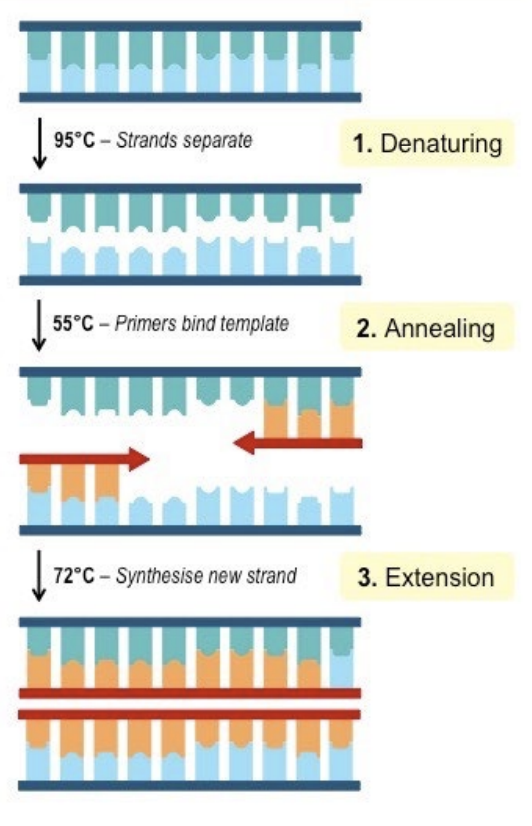
lysis
releasing of DNA from the cell
steps of DNA purification
DNA lysis
Purification of DNA from contaminants
*Successful DNA isolation requires methods that prevent nuclease degradation of the DNA
InstaGene Matrix
a commercially available kit to isolate genomic DNA, made with a specially formulated 6% w/v Chelex resin
bonds to PCR inhibitors, preventing DNA loss due to irreversible DNA binding
absorbs cell lysis products that interfere with the PCR amplification process
Polymerase Chain Reaction (PCR)
invented in 1938 by Kary Mullis, a scientist who worked for a biotech company based in the San Francisco area
valuable in DNA fingerprinting, detection of bacteria or viruses (particularly AIDS), and diagnosis of genetic disorders
What’s needed for PCR?
template: the DNA you want to amplify
Primers: short pieces of single-stranded DNA that are complementary to the target sequence
Nucleotides: (dNTPs or deoxynucleotide triphosphates; dATP, dCTP, dGTP, dTTP).
DNA polymerase: An enzyme that catalyzes the synthesis of new DNA strands by adding nucleotides to a template strand
The most common are Taq (from Thermis aquaticus), and Pfu DNA polymerase (from Pyrococcus furiosus).
both can generate new strands of DNA using a DNA template and primers, and are heat resistant
Cofactors: Magnesium ions needed for the proper functioning of the enzyme.
Thermal cycler: a machine programmed to alter the temperature of the reaction every few minutes to allow DNA denaturing and synthesis (see figure on next page).
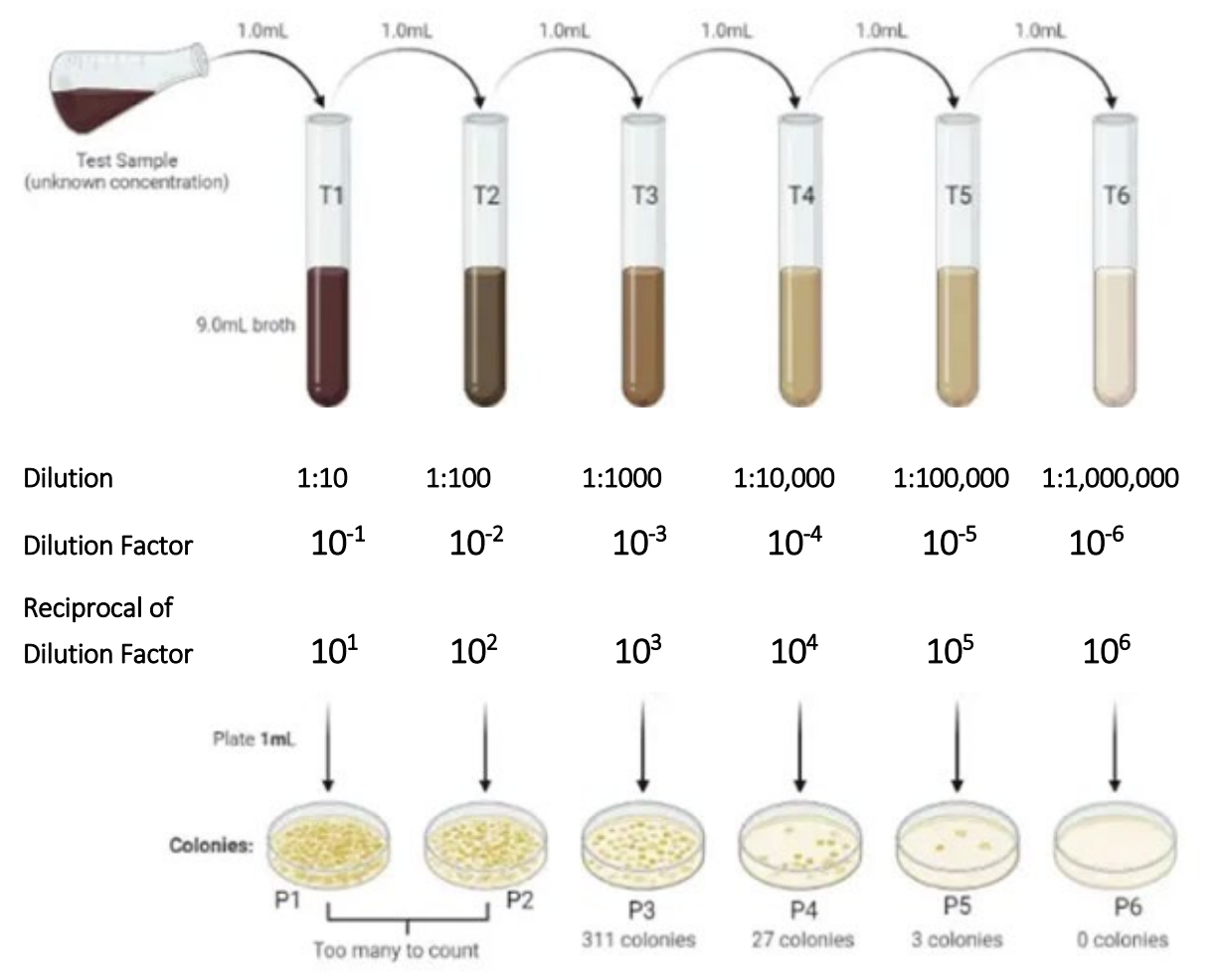
steps of PCR
denaturation → annealing → elongatio
denaturation
a segment of DNA is heated so it separates into two pieces of single-stranded DNA
annealing
the binding of primers to the single-stranded DNA templates
elongation
DNA polymerase uses the original strand of DNA as a template to add complimentary dNTPs to the 3’ ends of each primer and generate a section of double-stranded DNA in the region of the
product of PCR
The cycle of denaturing and synthesizing new DNA is repeated as many as 30 or 40 times, leading to more than one billion exact copies of the original DNA segment
limitations of PCR
highly sensitive: any contamination or trace amounts of DNA can produce misleading results
can only be used to identify the presence or absence of a known pathogen or gene
primers may anneal to DNA sequences similar to the target DNA
infrequently, incorrect nucleotides can be incorporated into the PCR sequence by the DNA polymerase
electrophoresis
the movement of charged particles in solution under the influence of an electric field
commonly used to separate nucleic acids or proteins on the basis of size, electrical charge, shape, and other physical properties.
agarose
a polysaccharide extracted from seaweed and used in gel electrophoresis
current travels:
cathode to anode
cathode
negative electrode
anode
positive electrode
DNA has a ___ charge at neutral pH
negative — which is why they move towards the positive charge in gel electrophoresis!
what size particles go farther in gel electrophoresis?
small particles move faster and farther, while large particles move more slowly and not as far
Agarose is almost sponge-like in composition, so it’s easier for smaller molecules to get through
loading dye
1) usually contains glycerol which will cause the DNA to sink in the well (rather than float away)
2) It contains a mixture of colored dyes which allow you to track the progress of the gel electrophoresis.
BLAST
Basic Local Alignment Search Tool — a suite of programs used to generate alignments between query and subject protein sequences
expect value
the number of times that an alignment is due to chance
Increased expect value = less reliable alignment (more likely due to chance)
reversion frequency
# revertant colonies per mL/ total CFUs/mL of overnight culture
chromosomal vs. extrachromosomal DNA
chromosomal:
contains essential genes
double stranded
linear or circular
found in nucleus or nucleoid
extrachromosomal:
Plasmid
circular, double stranded
contain non-essential beneficial genes for the cell (i.e., antibiotic resistance)
the boiling method
% agarose
determines the separation resolution
A higher percentage of agarose will resolve smaller DNA molecules and vice versa
true reversion
wild type → mutation by insertion → reversion via removal of the original insertion
second-site reversion
wild type → mutation by insertion → reversion by removal of another base
IFRM structure
implications, findings, results, and methodology
primary source
a source written/composed by the person/people/group who carried out the experiment(s) and/or research that will be explained in the piece