Molecular Evolution
1/29
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
30 Terms
Define evolution
Descent with modification from a common ancestor, leading to genetic and phenotypic changes over time.
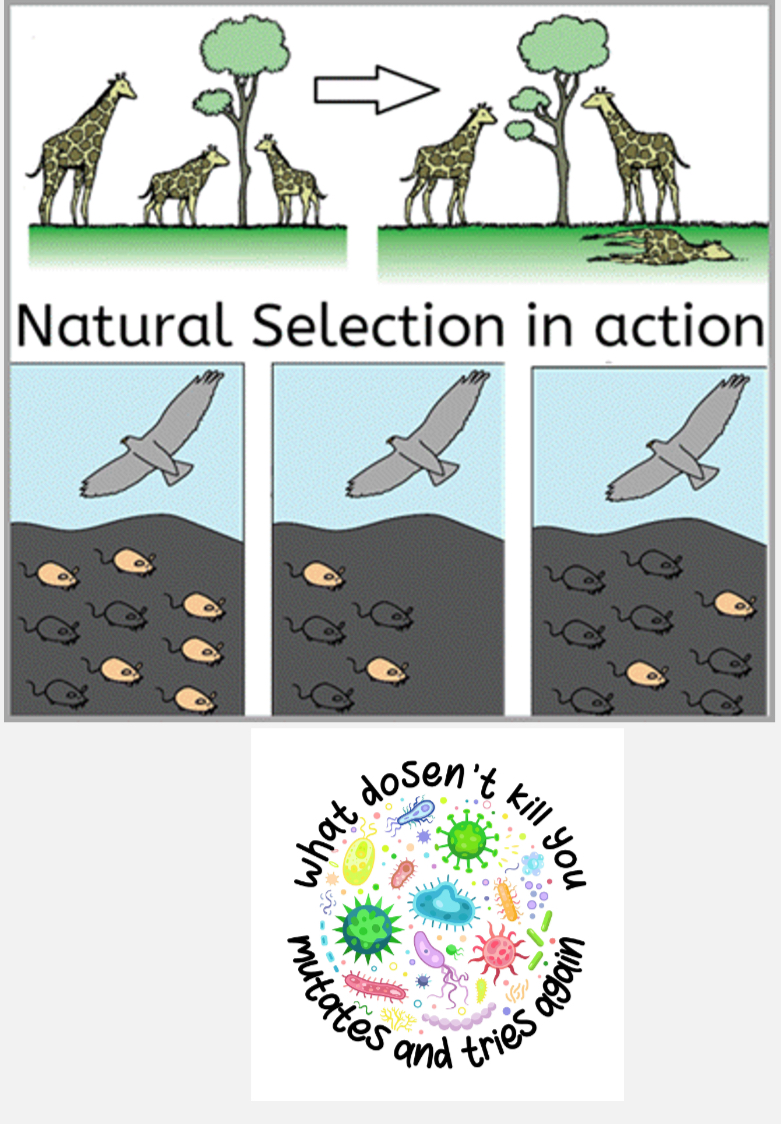
Natural selection
Process where variants that increase survival or reproduction become more common.
Fitness
The ability of an organism to survive and reproduce in its environment (often measured by reproductive success).
What is molecular evolution?
The study of how DNA, RNA, and protein sequences change over time, the forces shaping these changes (e.g., mutation, drift, migration), and how this impacts phenotype, function, and diversity.
What are the main mechanisms of molecular evolution?
Mutation
Selection
Genetic Drift
Gene Flow
Recombination
Gene Duplication
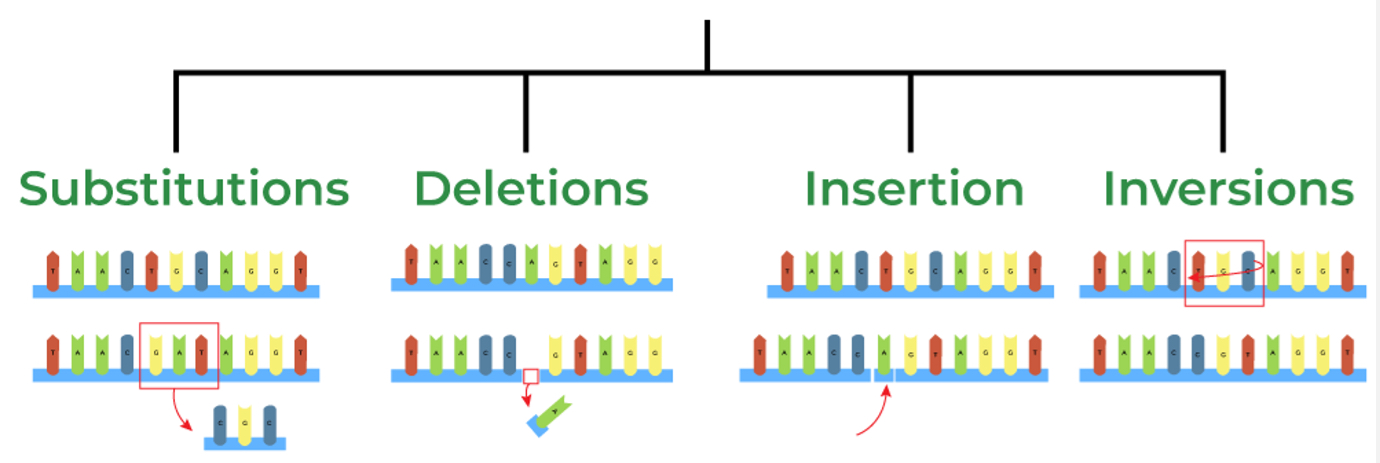
Mutation
Creates variation (errors, radiation, damage).
Selection
Non-random process favoring beneficial alleles.
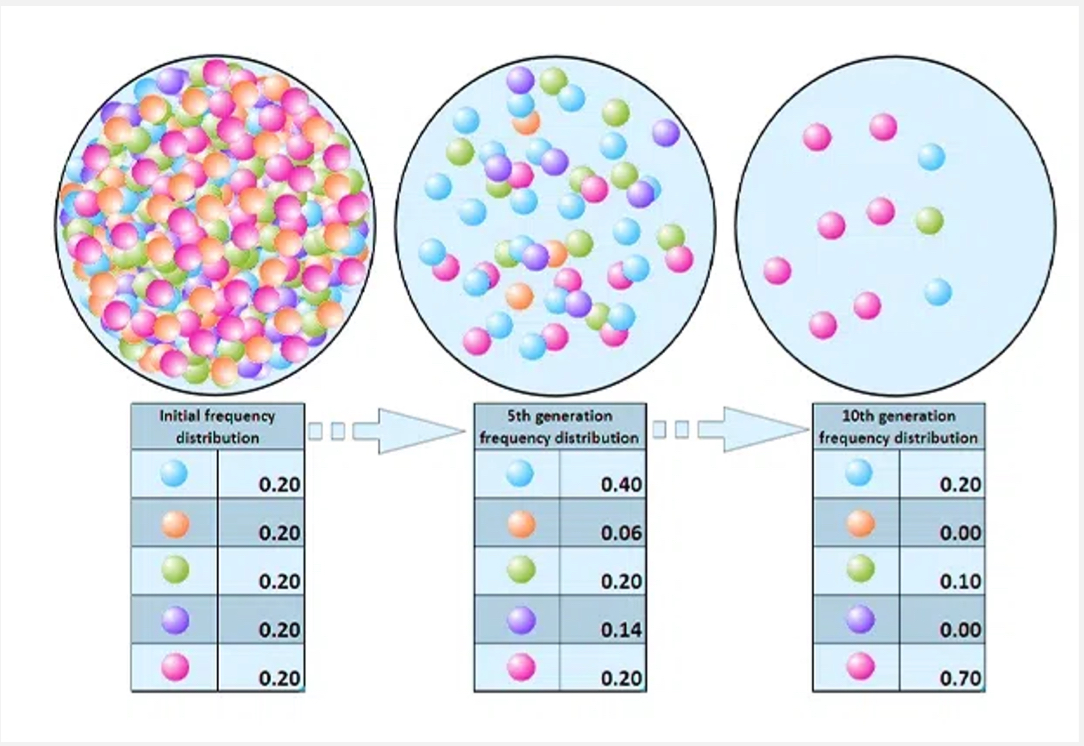
Genetic drift
Random fluctuations in allele frequencies (stronger in small populations).
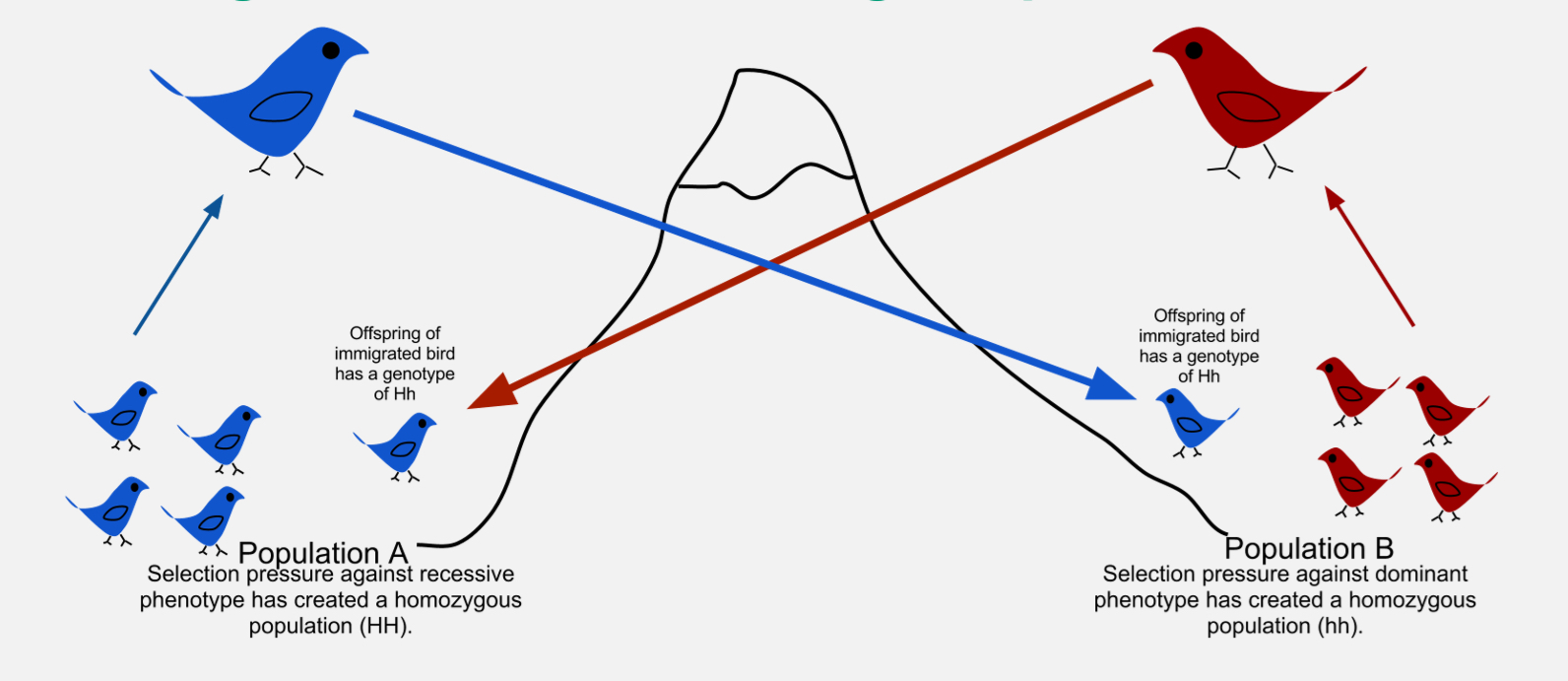
Gene flow (migration)
Movement of alleles between populations (can be adaptive, neutral, or maladaptive).
Recombination
Creates new allele combinations; may lead to duplication/deletion.
Reshuffles alleles during meiosis → creates new allele combinations.
Can bring together beneficial alleles or break apart harmful ones.
Mechanism for duplication and deletion (via unequal crossing over).
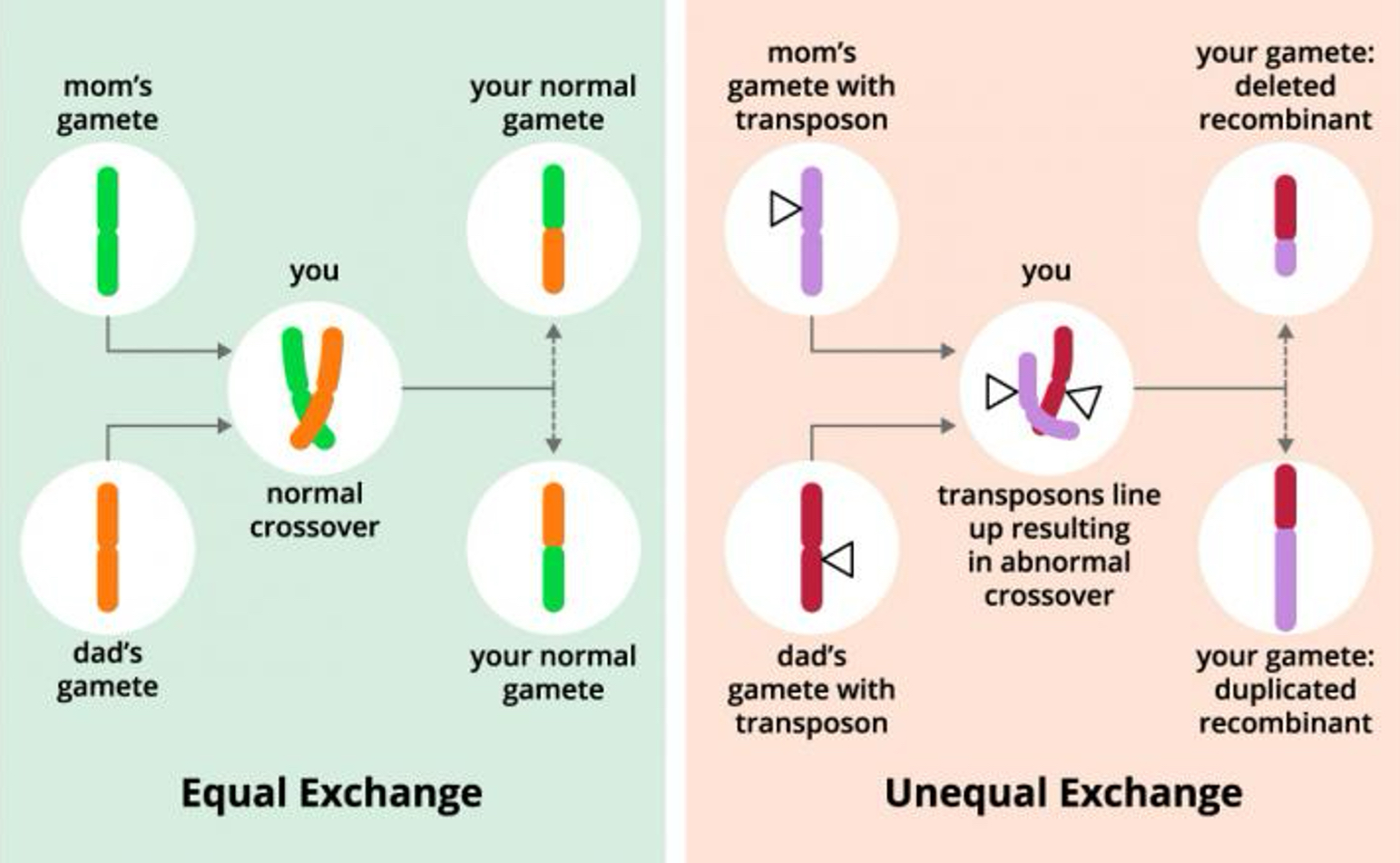
Gene duplication
Provides redundancy or new functions, leading to innovation.
Unequal crossing over during meiosis.
Retrotransposition (RNA copy reinserted into genome).
Whole-genome duplication (WGD) → entire set of chromosomes duplicated.
How do mutations drive evolution?
Introduce new alleles.
Lead to phenotypic variation (appearance, physiology, behavior).
Interact with the environment (selection acts on them).
Advantageous mutations may increase in frequency over generations.
What are the types of selection?
Positive selection
Negative/Purifying selection
Balancing selection
Sexual selection
Selected for = spreads
Selected against = fades out
Positive selection
Increases frequency of beneficial alleles (e.g., lactase persistence, CCR5Δ32 for HIV resistance).
Negative/Purifying selection
Removes harmful alleles (maintains conservation).
Balancing selection
Maintains diversity (e.g., heterozygote advantage in sickle cell).
Sexual selection
Traits improving mate attraction (e.g., peacock tails).
What is genetic drift and when is it most powerful?
Random changes in allele frequencies due to chance.
Strongest in small populations.
Can lead to fixation (allele becomes universal) or loss (allele disappears).
Examples:
Bottlenecks
The bottleneck effect occurs when a population undergoes a sudden, drastic reduction in size (e.g. due to a disaster or disease), leaving a small surviving population whose allele frequencies are not representative of the original population.
Caused by chance survival, not fitness
Leads to loss of genetic variation
Increases the impact of genetic drift, even if the population later recovers in size
Founder effect
The founder effect occurs when a small number of individuals break off from a larger population to establish a new population, carrying with them only a subset of the original genetic variation.
Allele frequencies in the new population depend on the founders
Rare alleles can become common by chance
Genetic diversity is usually low
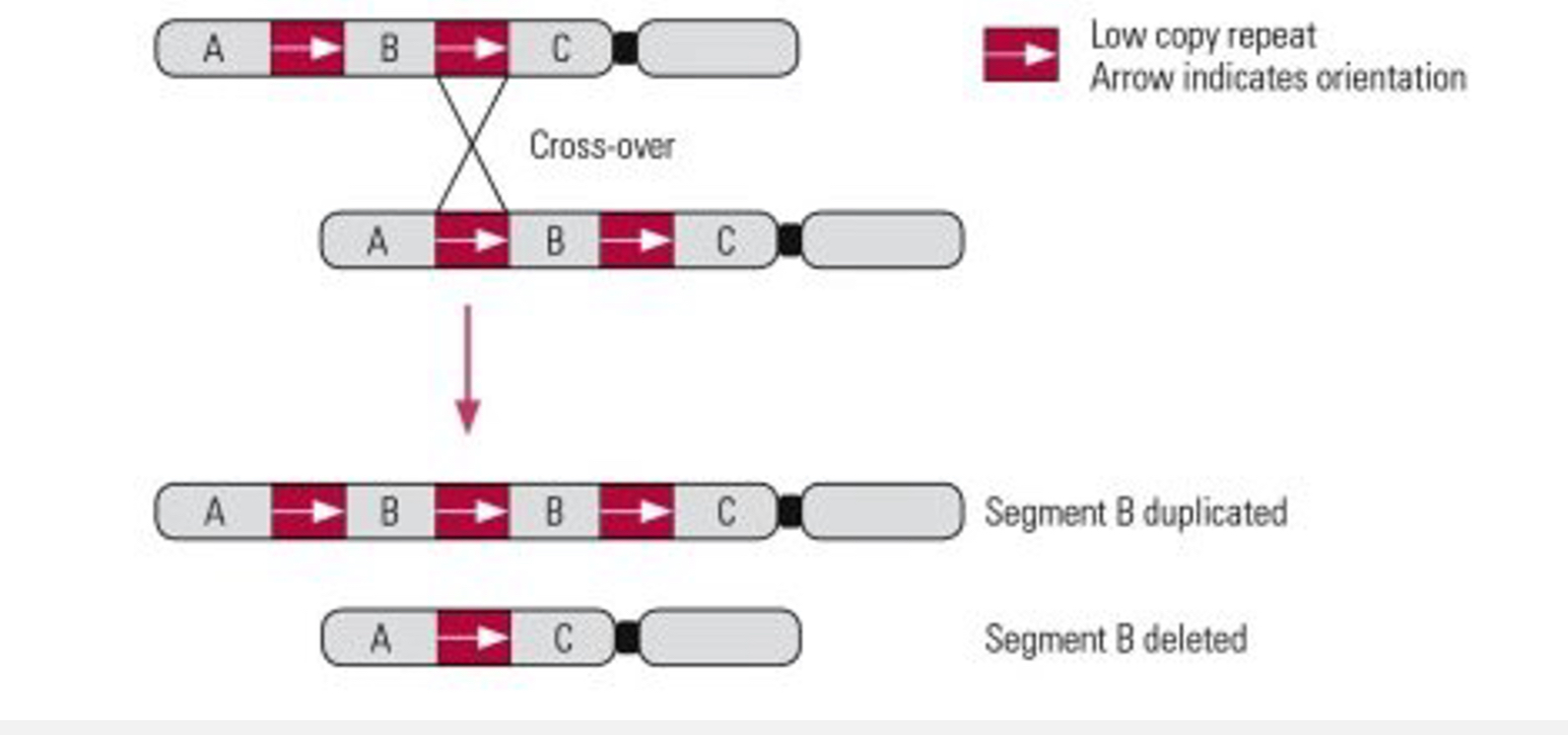
What is gene duplication and what happens after it?
Duplication of a DNA sequence provides evolutionary opportunities:
• Nonfunctionalisation: One copy becomes a pseudogene.
• Neofunctionalisation: One copy gains a new function.
• Subfunctionalisation: Functions split between gene copies.
Mechanisms:
Unequal crossing over
Retrotransposition
Whole-genome duplication.
What is sequence conservation?
When DNA sequences are highly preserved across species due to purifying selection.
Sequence conservation refers to DNA sequences that have remained similar or unchanged over evolutionary time because mutations in those regions are selected against (they would be harmful).
Harmful mutations do arise in important DNA regions
But purifying selection eliminates them
So over time, those regions appear conserved
Highly conserved: Coding exons, splice sites.
Intermediate: UTRs, promoters, terminators.
Low conservation: Introns, 3rd codon positions, intergenic regions.
Conservation = functional importance.
How is sequence conservation useful?
Identifying functional DNA/protein regions.
Predicting protein structure and function.
Locating regulatory sequences.
Understanding disease mechanisms.
Inferring evolutionary relationships.
Sequence Divergence
Sequence divergence refers to the accumulation of differences in DNA sequences between organisms or populations over evolutionary time. When two lineages split from a common ancestor, they initially have identical DNA sequences, but random mutations arise independently in each species over generations.
Some of these mutations persist, either because they are neutral or because they are favoured by positive selection, while others are removed by purifying selection if they are harmful. As time passes, the retained mutations cause the sequences to become increasingly different, leading to greater divergence. Regions under strong functional constraint show low divergence because mutations are selected against, whereas less constrained or adaptively evolving regions show higher divergence.
Causes: Mutations, natural selection, genetic drift, and reduced gene flow.
Measurement: Sequence divergence can be quantified by comparing nucleotide or amino acid differences.
Uses:
Helps estimate evolutionary time using molecular clocks.
Allows reconstruction of phylogenetic trees.
Reveals functional constraints (conserved vs divergent regions).
Explains how gene duplication leads to new functions (e.g., divergence of globin genes into fetal vs adult hemoglobin
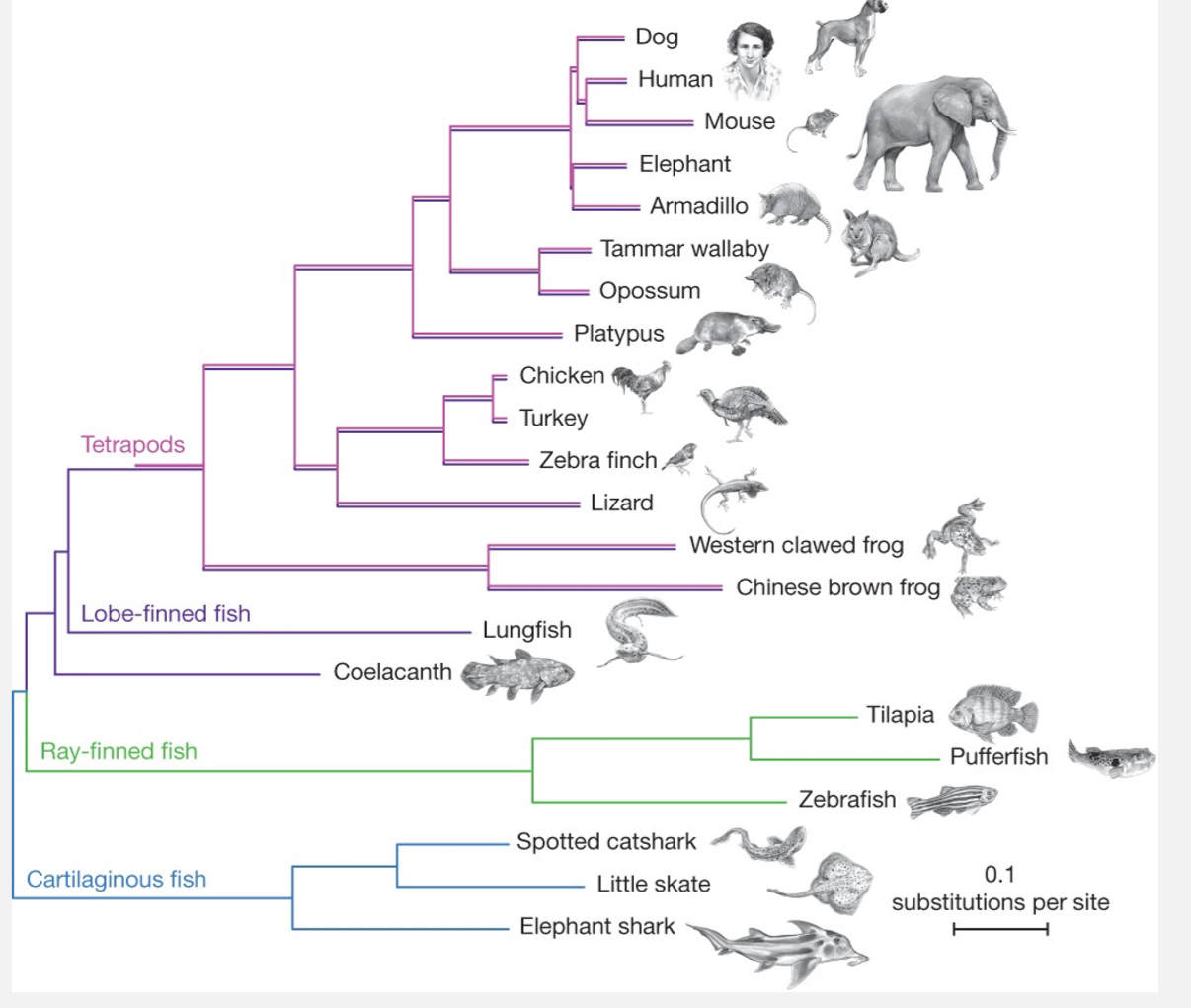
What is a phylogenetic tree and what is it used for?
A diagram showing evolutionary relationships between organisms, genes, or species.
Distance on the tree = degree of relatedness.
Built using mutation rates, selection pressure, and sequence comparisons.
Uses:
Reconstructing gene/species evolution.
Tracing disease origins (e.g., SARS-CoV-2 zoonotic origins).
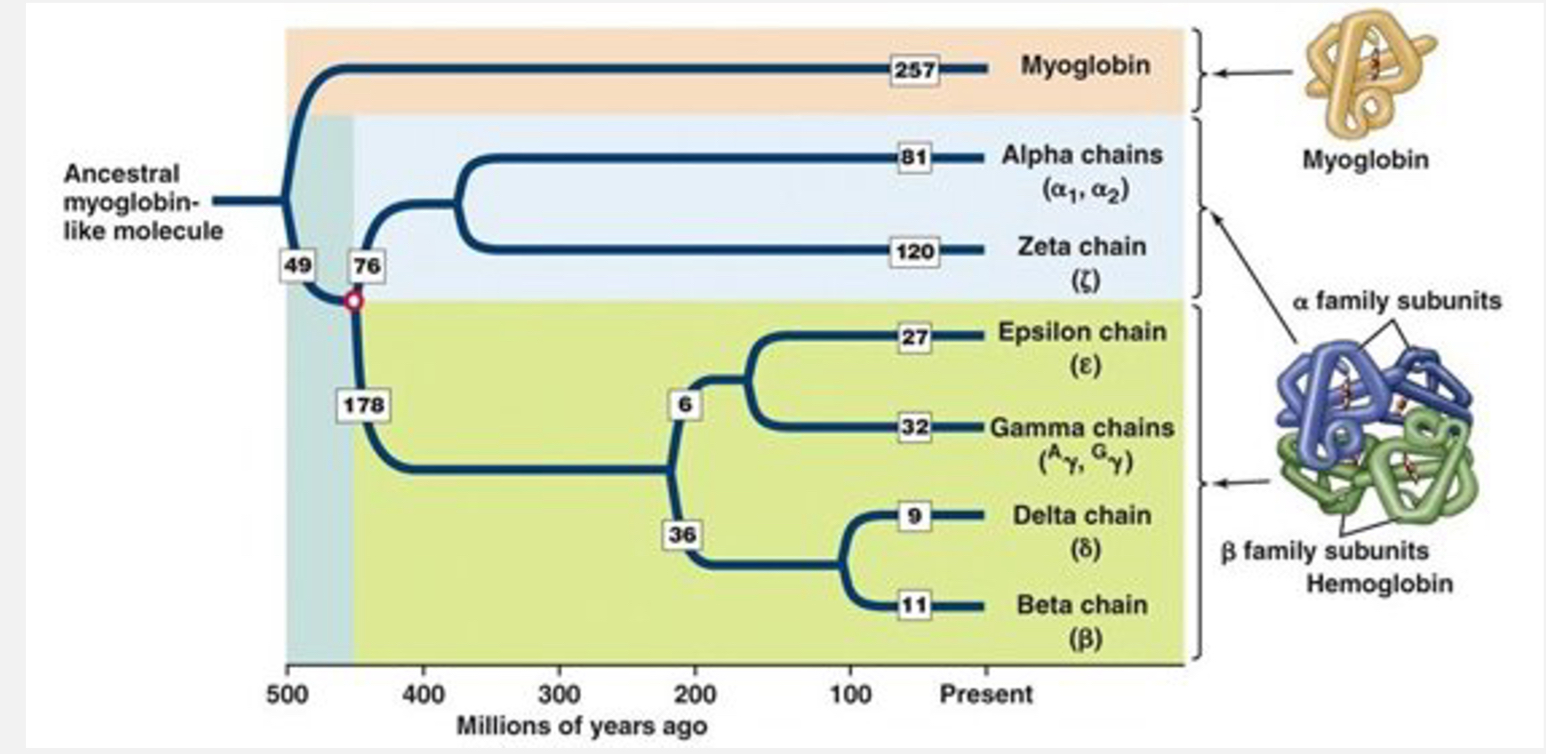
What is the globin gene family and why is it important in molecular evolution?
Originated from: ancestral duplication which split into myoglobin + hemoglobin → α-like and β-like clusters.
The globin gene family arose from a single ancestral globin gene that underwent repeated gene duplication events during evolution. After duplication, the copies accumulated mutations and diverged slightly, becoming paralogous genes with related but specialised functions. Some duplications occurred locally along the chromosome, producing clusters of globin genes, while earlier whole-genome duplications helped separate α-like and β-like globins.
Through a process called subfunctionalisation, different globin genes became optimally expressed at different stages of development—embryonic, fetal, and adult—allowing efficient oxygen transport under changing physiological conditions. Natural selection retained these duplicated genes because their complementary, stage-specific roles increased fitness, while non-functional copies became pseudogenes.
Stage-specific expression: Embryonic → foetal → adult globins.
Paralogs retained: By subfunctionalisation and natural selection.
Includes pseudogenes as evolutionary relics.
Shows interplay of duplication, divergence, and selection.
How do haemoglobins change during development?
Embryo: HBZ, HBE1 → high O₂ affinity.
Foetus: HBA1/2, HBG1/2 → pull O₂ from maternal blood (normally silenced after birth).
Adult: HBB, HBD → optimized for tissue O₂ delivery (mutations here clinically important).
What causes Sickle Cell Disease (SCD)?
SNV in HBB gene: Glu6Val mutation.
Homozygous (HbSS) → SCD.
Heterozygous (HbAS) → carrier, usually healthy, resistant to severe malaria.
Mutation ~7300 years ago.
Common in malaria-endemic regions (Africa, Middle East, Mediterranean, India).
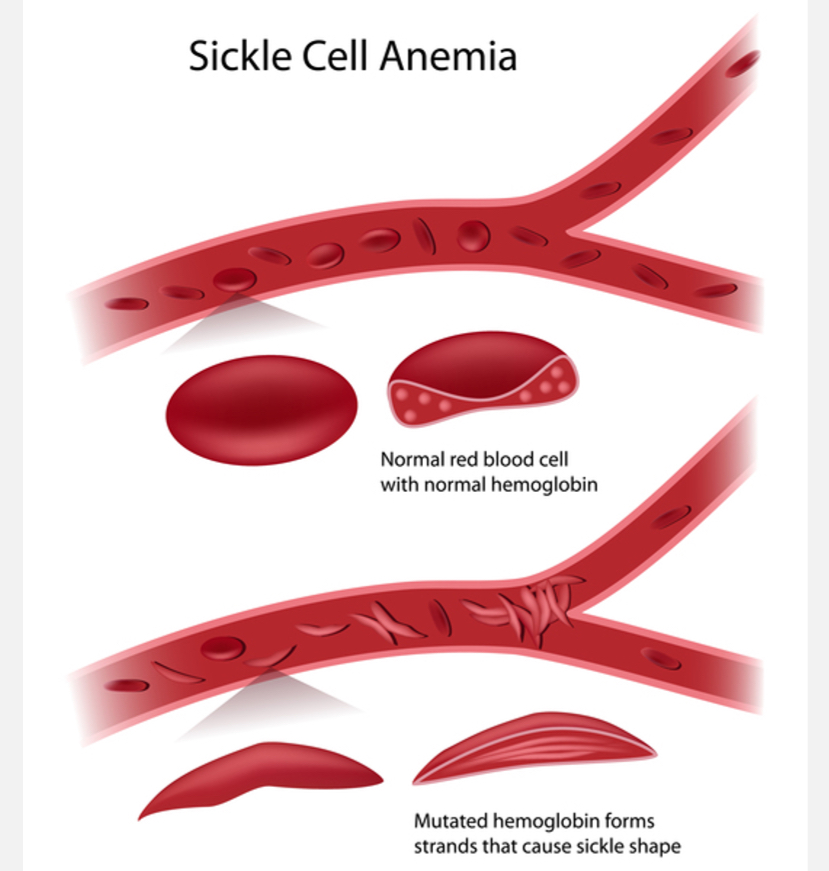
What is the link between sickle cell and heterozygote advantage?
HbAA: Healthy, but malaria vulnerable.
HbSS: Sickle cell disease, reduced survival.
HbAS: Carrier, healthy + malaria-resistant.
This balancing selection maintains the HbS allele in populations → classic case of evolution shaping human health.
Heterozygote advantage: Where individuals carry 2 different alleles for a gene have a higher fitness(survival) than individuals who are homozygous for either allele
What is a pseudogene?
A DNA sequence resembling a gene but nonfunctional due to mutations or lack of regulatory elements. Often arises from gene duplication and nonfunctionalisation.
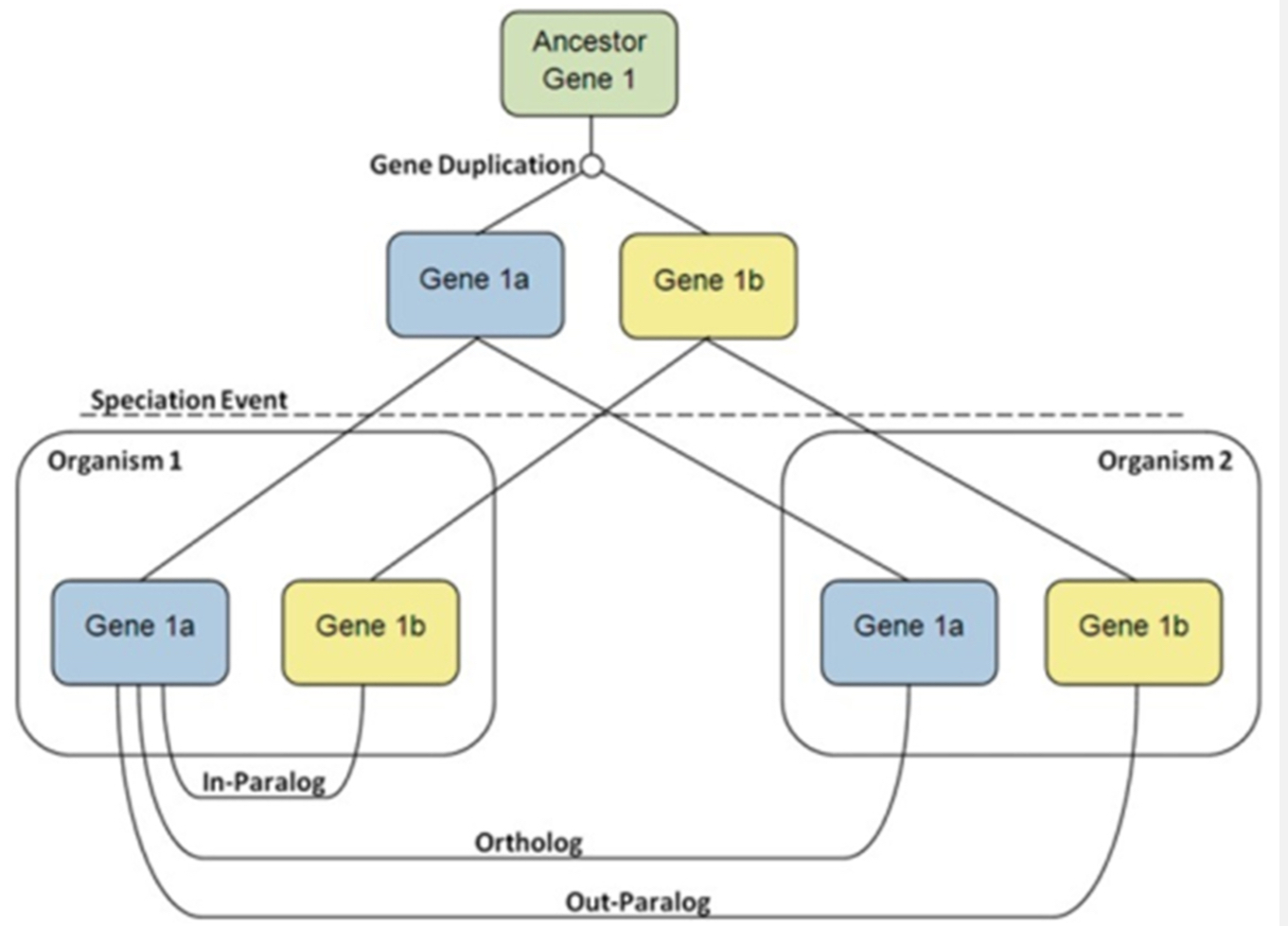
What are homologs, orthologs, and paralogs?
Homologs: Genes related by descent from a common ancestor.
Orthologs: Same gene in different species, diverged by speciation (e.g., human HBB and mouse Hbb).
Paralogs: Duplicated genes in the same genome (e.g., human HBA and HBB).
Give examples of positive selection in humans.
Lactase persistence (LP): continued ability to digest lactose in adulthood.
CCR5-Δ32 mutation: confers resistance to HIV infection.
Antibiotic resistance in bacteria.