W5: Transposon tools for insertional mutagenesis and targeted expression
1/39
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
40 Terms
Transposable elements
‘transposons’
account for most dispersed repetitive DNA in eukaryotic genomes
BUT ALSO
really useful in the lab!
Why useful in the lab?
Mutagenesis
Insertional mutagenesis
Ease of crossing
Enhancer trapping
Mutagenesis→ why useful
large scale unbiased mutant screens→ great for dissecting mechanisms of cell function and development
Generate loads of random mutations in genome
screen for occasional mutations that affect process of interest
e.g cell division, embyronic development, cell death, behaviour etc
any mutant that affects this must have hit important component of process
Advantages of this approach
no need for preconceptions about
nature of protein, cellular location, enzymatic properties
Only need informative phenotype
Disadvantages of this approach
Following up interesting mutations needs→ cloning of gene
route to molecular analysis of its function
e.g If used Chemical metagenesis
generates many point mutations
and affected gene must be identified by ‘positional cloning’
easier than it used to be but not trivial
Insertional mutagenesis (compared to chemical mutagenesis→ why better approach!)
mutation caused by insertion of DNA fragment into gene
this fragment carried a molecular ‘tag’
can be easily identified by mutant phenotype
much easier to clone and analyse molecularly
→ than a gene in which only point mutations are available
Ease of crossing of these mutants
if transposon carriers a dominant marker
(e.g white+→ expressed from its own promoter)
→ it is easy to follow the insertion in genetic crosses
If carried a recessive marker
e.g mutant phenotype caused by the insertion
→ not as easy!
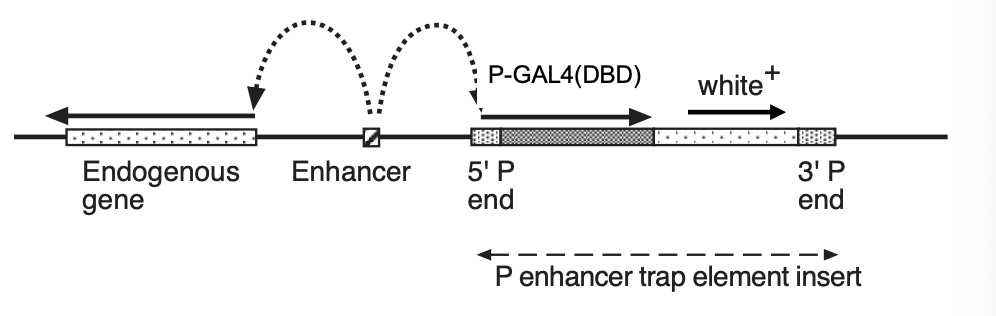
Enhancer trapping
Consider:
transposon construct with reported gene fused to minimal promoter
e.g→ GAL4(DBD): encodes DNA binding domain (DBD) of the yeast transcription factors GAL4
significant transcription only occurs when the element integrates next to the enhancer in the genome (figure)
As most enhancers are cell-type specific
→ different insertion sites of an ‘enhancer -trap’ element therefore allow many cell types to be labeled by GAL4 expression
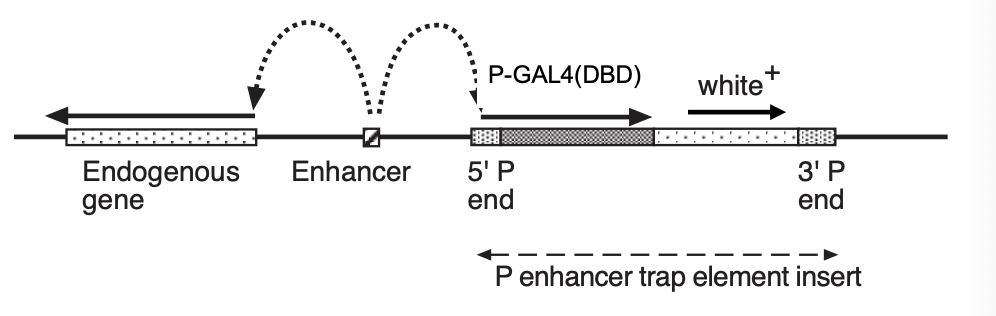
It may also be possible to identify…
a nearby gene that shows the same expression pattern as the GAL4 reporter
why:
because it is driven by the same enhancers
In order for transposons to be useful in the lab
need transient controlled transposition
so mobile DNA can insert at new genomic locations at a high enough frequency
Drosophila P transposon
encodes Transposase
→ needed for tranposition of the element
Transposase binds to specific sequences near end of P element
together with endogenous Drosophila proteins→ catalyses
excision of element
and
sometimes transposition
Therefore: P transposon will always EXCISE element→ but not always put it in somewhere else!!
Partially deleted P element without transposase can still transpose if
There is another source of transposase
it has cis-acting sequences
that are necessary to bind transposase
has other necessary host proteins
A single intact transposase gene anywhere in the genome can…
Catalyse transposition→ in trans
of any other P element which has all the needed cis-acting sequences
A partially deleted P element which lacks intact transposase gene (but with all needed cis-acting sequences for transposition) will…
remain stably integrated→ in the absence of transposase
will NOT tranpose further
Using these principles in the practical
Used to recover new insertion of a P Element
Element used:
enhancer tap construct
THEREFORE
will also assess whether any new insertions recovered are giving novel patterns of GAL4 gene expression
Thus→ potentially detecting novel cell types and genes and generating genetic tools to manipulate these cells
Plus acquiring and optimising fluorescence microscopy images
Actual Experiment stuff:
How to identify new P element insertions
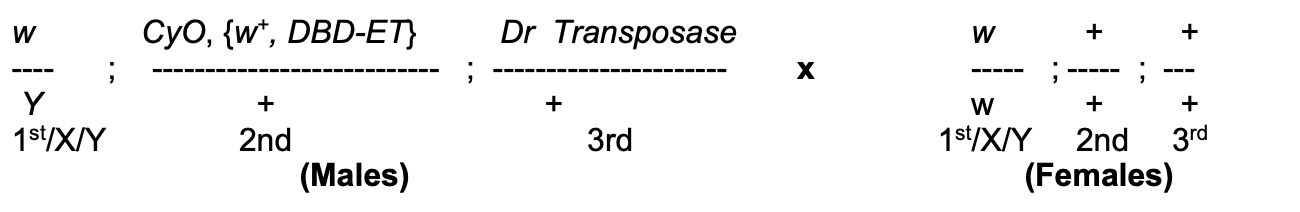
Experimental background→ cross of flies set up
Females→ wild types
Males→ carry
P construct P{w+, DBD-ET}
carries white+ gene inserted on a 2nd chromosome (CyO→ Curly of Oster)
which is marked by Curly (Cy)
Mutation in endogenous w gene on X
THEREFORE: only w+ is on the P element
(so can follow the P element)
CyO→ ‘balancer’ chromosome
carries multiple inversions which when heterozygous with wild-types chromosome→ prevent recovery of recombinants in the progeny
HOWEVER: (not needed just yet) coz male Drosophila=no crossing over
but still needed for females in other parts of crossing scheme
Third chromosome→ with tightly linked (so make sure know where transposase is)
P transposase gene
and dominant marker→ Dr (Drop)
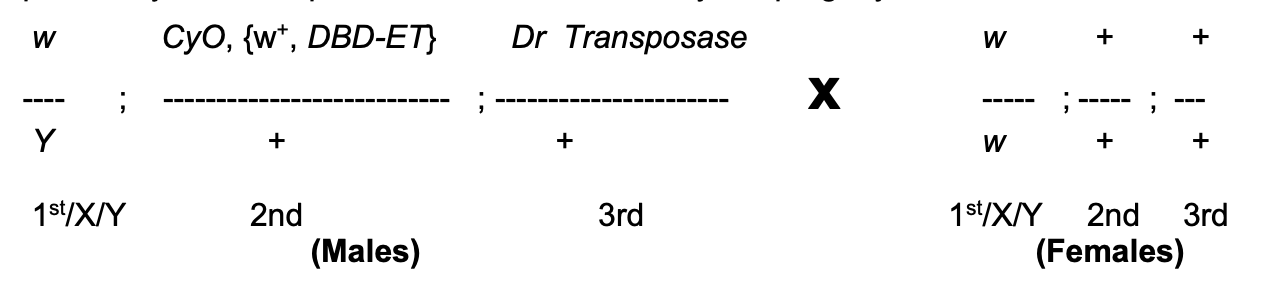
What these modifications to the male fly allow
Mobilisation of P elements in the male parents
If happens in germline cells→ progeny could contain new P insertion (or just excision) in all of their cells
If transpoase expressed in somatic cells→ still some insertion/excision BUT not transmitted to progeny
(so not interested)
THEREFORE: by studying the progeny of this cross→ can see what these new insertions/excisions look like!
What are the next steps in this practical
Identify individual flies that carry new P insertions→ new transposants
Cross these to partners (with flouresencne gene)
Obtain larva to examin for expression of GAL4(DBD) enhancer trap
Deciding how to identify new transposants: Questions to ask
Q1a. Work out the genotypes of the gametes produced by each parent in the above cross
Q1b. Work out all the classes of progeny - their full genotypes and their phenotypes (including their sex) - of the above cross. A Punnett Square may help you to do this. For now, don't (yet) worry how transposition may affect this.
Q2. Your next task is to work out how to spot occasional flies that MUST carry a new insertion. To work towards this:
Q2a. Should you keep or discard flies with the original P insertion? How can you do this?
Q2b. Should you keep or discard flies expressing transposase? Why? How can you do this?
Q2c What flies will carry neither the original P insert, a new P insert, nor transposase?
Q2d. Finally, which flies (the ones that you want) carry neither the original insert nor transposase, but do carry a new insert?
P.S. Does it matter whether you select males or females?
Q1a: work out genotypes of gametes produced by each parent in the cross
Just mix all the ways to combine together with punnet square
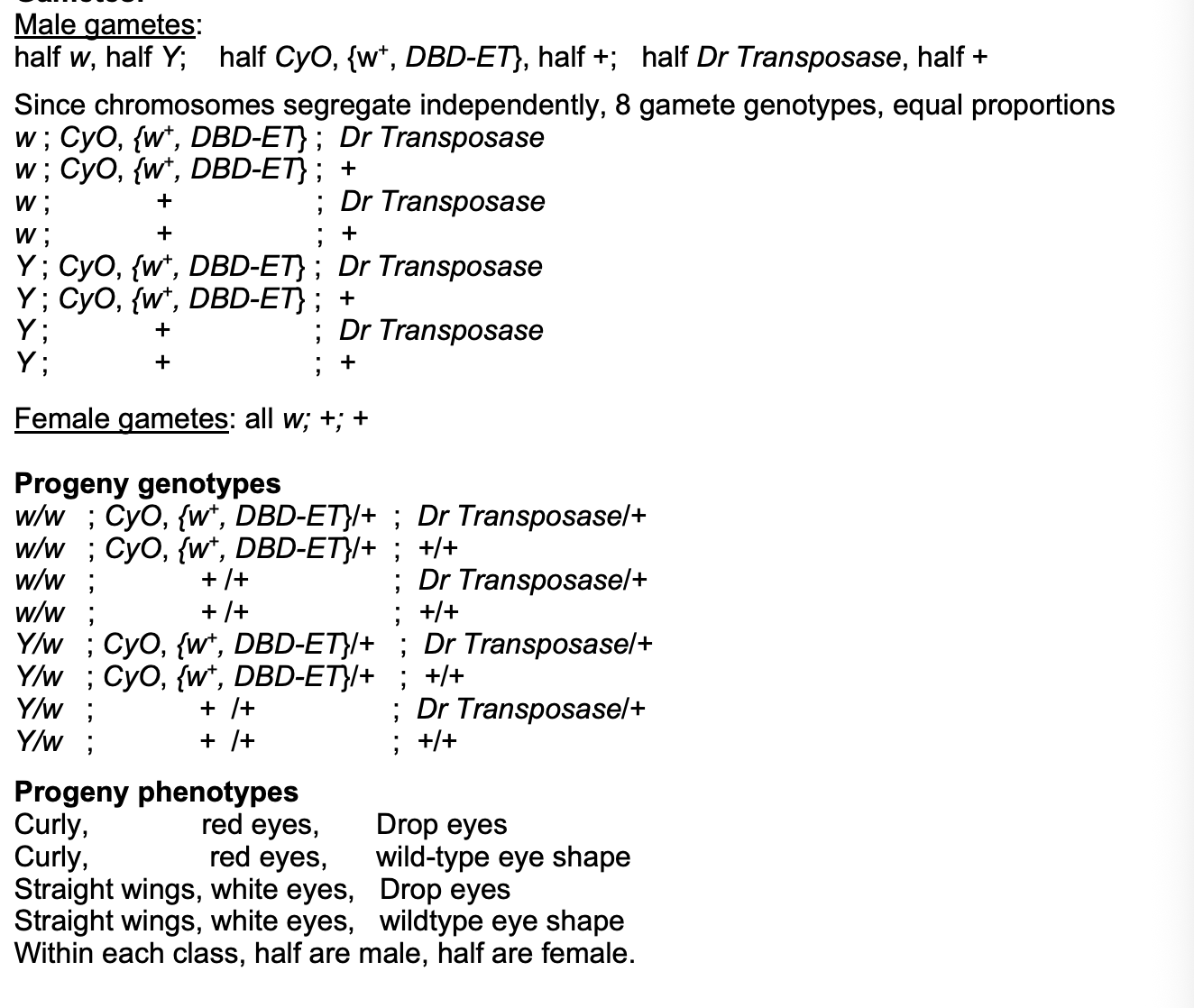
Q1b: Work out all the classes of progeny
Progeny phenotypes
Curly, red, drop
Curly, red, WT
WT, white, Drop
WT, white, WT
Half male and half female
Q2a: Should you keep or discard flies that carry the original P insertion? How to do this?
If progeny:
Curly→ will carry P{w+} too
means has original insertion and NOT a new one (not of interest!)
→ IGNORE: curly red-eyed
What to look for: flies with non-curly wings
Q2b. Should you keep or discard flies expressing transposase?
DO NOT WANT
Why:
we want to keep the new insertions made stably integrated and non-mobile
if tranposase is still expressed→ new insertions might be lost/move again
HOW TO GET RID
Keep only Dr+ flies
Coz Dr marker is on transposase-producing chromosome
Q2c. What flies will carry neither the original P insert, a new P insert, nor transposase?
white-eye, non-curly which are not Dr
THEREFORE: get rid
Q2d. Which flies carry neither original P insert nor transposase, but carry a new P insert?
Red eyes, non curly, not Dr
What is the genotype of the vigin females
w; +; UAS-GFP, elav-AD
homozygous for UAS-GFP and elav-AD constructs
→ (homologous chromsome pairs are separated by semicolons)
This will provide the florescence for visualistion of this progeny
Q3: Could different transposant individuals carry identical or non-identical insertions? For this answer, does it matter whether they came from different vials or the same vial? Hint: does the stage of development when transposition occurs matter?
DIFFERENT VIALS: New transposants from different vials are very unlikely to carry insertions at the same location
why: P element can integrate throughout the genome
SAME PARENTAL VIAL:
may sometimes also applies
BUT OFTEN: carry the same new insertion
WHEN: if transposition occurred early enough in the germine for mitotic cell division to have produced many germ cells (with the same insertions)
THEREFORE: depends on timing of the insertion event if all have same insertion in the same vial
4. Could mobilisation occur in somatic cells of the male parents? Does this matter?
transposase is expected to express in somatic cells of the male parent
but
this will have no influence on classes of progeny obtained
coz somatic cells do not contribute to gametes
5. When the progeny of your crosses hatch as adults, what proportion of the progeny do you expect to carry an insertion?
Progeny carry a 1:1 ratio of new insertion to no insertion
coz ust a 50:50 chance
The most common event is single insertion of P{w+, DBD-ET}
which is heterozygous
THERFORE: 50:50 chance
6. Why will some insertion events be mutagenic and others not? Why will insertion into a gene normally cause a recessive mutant phenotype?
some insertion events will be in genes (or their regulatory sequences)
→ THEREFORE: mutant genotypes
others will be in intergenic regions which will
not inactivation of any gene→ NO phenotype
some will inactivate genes not necessary for viability→ cause phenotype that is not immediately obsvious to a superfial inspection
→ YES PHENOTYPE BUT NOT SEEN (that is really seen)
Insertion of P element will probably inactivate the gene
supplying a copy of the wild-type gene will usually supply enough gene product to produce WT phenotype
→ THEREFORE just recessive
7. Sometimes mobilisation excises the P insertion. Some excisions appear “precise”, some leave some P element sequences present, and some excise flanking genomic DNA.
How could you identify any progeny that carry such excisions?
Why might excision of flanking genomic DNA sometimes be useful for us?
White-eyed, Curly flies
lost the w+ but still have curly CHECK THIS
Excisions of flanking DNA may sometimes generate useful delections of nearby genes
How could you establish a stable stock of any new insertions? Think: How can you maintain a stock of an insertion that is homozygous lethal?
Principle→ from existing knowledge:
simplest stable sock is one homozygous for the insertion
How to get this?
crossing two heterozygous parents for the same insertion
BUT: initial transposant male fly (red-eyed non-curly non-Drop male_ cannot e crossed to itself
THEREFORE: must first cross it to generate F1 flies
HALF: will be heterozygous
THEN cross these heterozygous males and females to eachother
Must need a way to identify heterozygous and homozygotes
Way to identify heterozygous and homozygotes
Need suitable genetic markers
Provided by balancers→ e.g CyO for insertion on a second chromosome
For this example of 2nd-chromosome insertion:
cross heterozygous w+ insertion fly x w- balancer stock (w;CyO/If)
If→ dominant eye shape marker
THEREFORE
in F1→ half will be heterozygous for the w+ insertion and half will carry no insertion
of w+ F1 flies→ half will also carry CyO and be {w+}/CyO
Cross these heterozygous males and virgin females to each other
25% of their progeny will be homozygous
→ can be identified as non-curly
How can you maintain a stock of an insertion that is homozygous lethal?
If it is maintained heterozygous with different homozygous lethal mutation
→ only heterozygotes are viable
But this only works indefinitely if you…
suppress the recombination between the two homologous chromosomes
→ need to prevent recombination giving rise to homozygous viable recombinants that carry neither the lethal insertion nor the homozygous lethal mutation
How can this be done?
having your insertion heterozygous with a balancer chromosome
→ which also carries a homozygous lethal mutation (or mutliple)
Procedure of W5 practical
Given 3-4 vials of progeny from fist cross, need to
Check you are finding all classes of progeny that you expect to find
Screen for any flies with new insertions
set up crosses as described, we provided partners
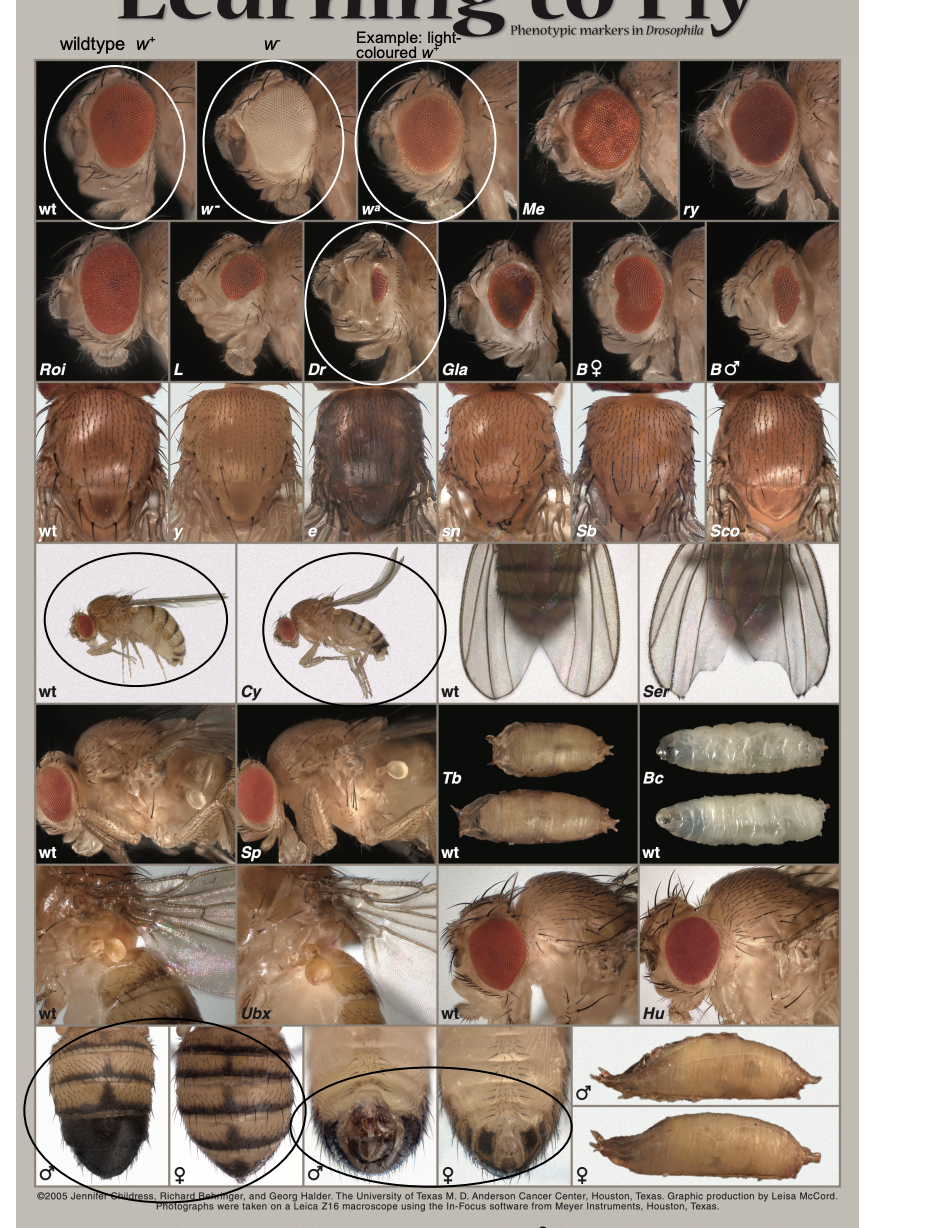
Recognising phenotypes and markers
Eye colour markers
w+- red, w- white
w+ inserts→ may only be pale red, yellow, orange
not always expressed at high enough level for WT
2nd Chromosome markers
Curly→ dominant wing phenotype
BEWARE: can be mild and hard to distinguish from WT
3rd Chromosome markers
Dr (Drop)→ greatly reduced eye
easily scored BUT don’t mistake lack of normal-sized red eye for w-
Sexing flies
Male→
black patch on the dorsal posterior abdominal segments
one segment fewer than females
sex combs on the front legs of the males (reliable indicator)
USE microscopes for this to make sure!
Things to be cautious of
do not over-anaesthetise→ use minimum CO2
do not allow to stick onto the food
Do not bang tube on bench→ might bang food out
hold together and tap hands gently onto the bench
Dispose of unwanted flies into morgue asap→ avoid cross-contamination
Each cross wit single new insertion (enough just one insertion→ see above) and note if from same or different vial
Use virgin females→ because females will store the sperm