BIOL 2030: Module 12
1/53
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
54 Terms
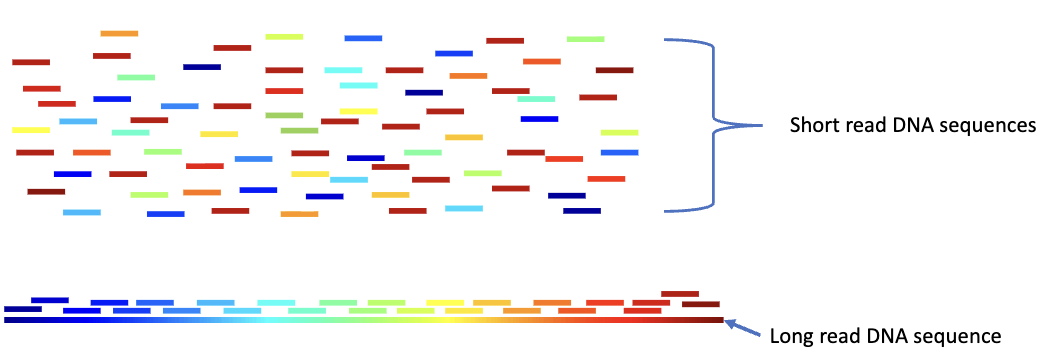
Shotgun Sequencing
shear genomic DNA into short sequences
sequences by next gen.
assembler software looks for sequence overlaps between fragments to assemble them into larger fragments (contigs)
now preferred way of sequencing genomes, but has problems with repetitive DNA sequences
long read sequences often used to overcome this problem
genome assembly by shotgun sequencing is a big bioinformatic task
each segment is sequenced on average 100s of 1000s of times
Long Read Sequences
help with the assembly and alignment of short reads
ex. Nanopore
Read Depth
how many times a given DNA base is sequenced in an experiment.
10x read depth means each base was sequenced 10 times
greater read depth gives more confidence a base is accurately read
initial read depth is usually several hundreds to thousands
when additional copies are sequenced, much lower read depths are sufficient
Base Calling
the crucial step in DNA/RNA sequencing where raw data (like electrical signals or light intensities) from a sequencer is translated by software into the actual sequence of nucleotide bases
greater depth gives more confidence a base is accurately read
Shotgun Sequencing Application
transcriptomics via next-gen
gene expression analysis
indentifying species (DNA barcoding)
studying microbiomes via next gen
environmental DNA via next gen
Transcriptomics
the study of the complete set of all RNA molecules (transcripts) in a cell or population of cells at a specific time, representing a snapshot of active gene expression
The number of times each gene appears in the shotgun sequence data is a measure of the degree
to which that gene was being expressed in the organism/tissue being studied
Identifying Species via DNA Barcoding
though many genes/DNA sequences options, mDNA COI gene is the most widely used for identifying animal species
other genes are used for plants and fungi
example: using DNA barcoding to identify sea food, or uncovering the evolutionary relationships of living and extinct species of humans
Studying Microbiomes (next gen)
how its done:
isolate DNA from an environmental sample
amplify microbial sequences using primers that amplify 16 rDNA gene
sequence using next gen (ex. illumina)
run data through databases to see what species are present, and in what relative abundance
Environmental DNA (next gen and qPCR)
DNA can be isolated from environmental samples
even in air samples
using appropriate primers, informative DNA sequences can be amplified, and then sequenced.
species that are present can be identified via use of species/DNA databases
particular species can also be targeted using taxon-specific primers, followed by qPCR
Why Study Genetic Variation
to determine the genetic basis of inherited diseases or phenotypic traits
to study the relatedness of individuals or populations, and degree of intermixing of populations
to identify individuals
parentage analysis
to identify criminals
DNA Fingerprinting
invented using minisatellite DNA
consist of 10-100 bp sequences that are repeated many times in tandem arrays
minisatellite DNA loci have extremely high allelic variation, due to frequent mutatiosn involving replication slippage errors and/or unequal crossing over
originally relied on Southern blotting, a tedious method no longer used
now done using microsatellites (STRs and SSRs) which have shorter sequence repeats than minisatellite DNA. .
can be amplified via PCR
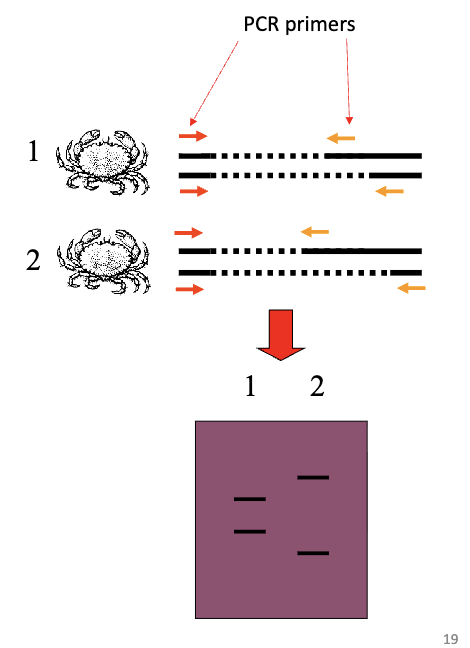
Microsatellite Genotyping
PCR primers designed for flanking sequences
primers are fluorescently labeled, and amplify products of different size
separate products by electrophoresis
genotypes identified by size of products
co-dominant
Co-dominant
heterozygotes produce 2 bands
both alleles are detected
Microsatellite Genotyping Equipment
usually use the same capillary electrophoresis machines used for dideoxy sequencing
multiple microsatellites often amplified at the same time, using primers labelled in different fluorescent colours
multiplex analysis
Microsatellite DNA in Forensics
13 standard microsatellite loci are enough to distinguish all human individuals
only requires a small amount of DNA
contamination is often a problem due to sensitive methods
Trinucleotide Repeats
in humans, a few microsatellites cause disease. in all cases, these loci invovle trinucleotide repeats within genes, or other important DNA sequences
all humans have these microsatellite loci, however healthy humans have alleles with a small number of repeats.
humans with genetic disorders have versions with too many repeats. these versions cause production of abnormal proteins
cause of”
Huntington's
Fragile X
Myotonic Dystroph
RFLP Analysis
restriction fragment length polymorphism
use of restriction enzymes to detect DNA polymorphism
mutations can either create or destroy restriction endonuclease sites
gain or loss of restriction sites can be detected using gel electrophoresis
restriction site polymorphisms are most commonly caused by single nucleotide polymorphisms (SNPs)
Single Nucleotide Polymorphism (SNPs)
SNPs caused by single base mutations are the most common genetic variations in genomes
SNP occurs every 800-1000 bp in human DNA
any 2 humans have different SNP alleles at several million SNP loci
the average genome differs from standard ‘reference’ genome
polymorphism is usually di-allelic
SNPs close to each other on a chromosome are usually inherited together, forming haplotypes
Diallelic Polymorphism
a common genetic variation where a specific DNA site (locus) has exactly two common forms (alleles) in a population
due to either:
SNP
indel
Haplotype
arbitrary long stretch of DNA characterized by particular alleles at the SNP positions in that sequence
physically close
usually inherited together
SNP Chips
used to genotype large numbers of SNPs
utilise DNA hybridization-based assays to determine genotypes at known SNPs
have become the general method of choice for rapidly screening millions of loci at once
GWAS
genome wide association
aim is to find genetic links to diseases
look for SNPs that have alleles correlated with presence for disease/trait
some diseases/traits are entirely or mostly determined by a single gene (ex. cystic fibrosis, sickle cell anemia, ear wax composition)
need to survey many SNPs, and many individuals
CRISPR-CAS
bacterial defence against foreign DNA now used by molecular biologists as a genetic engineering tool
interest in CRISPR-Cas9 type
Clustered Regularly Inter Spaced Palindromic Repeats
CRSPR ASsociated proteins
designed to target specific DNA molecules, comparable to adaptive immune systems of vertebrates
How CRISPR Works
Targeting
Binding
Cleavage
DNA Repair

CRISPR Targeting
Scientists introduce the Cas9-guide RNA complex into a cell where it randomly associates and dissociates with the DNA. Cas9 recognizes and binds to PAM
CRISPR Binding
Once it binds to a PAM motif, Cas9 unwinds the DNA double helix. If the DNA at that location perfectly matches a sequence of about 20 nucleotides within the guide RNA, the DNA and matching RNA will bind through complementary base pairing
CRISPR Cleavage
The DNA-RNA pairing triggers Cas9 to change its three-dimensional structure and activates its nuclease activity. Cas9 cleaves both DNA strands at a site upstream of PAM.
CRISPR DNA Repair
Cells contain enzymes that repair double-stranded DNA breaks. The repair process is naturally error-prone and will lead to mutations that may inactivate a gene. Cleaving DNA at a precise location is one of many applications of the CRISPR-Cas9 technology.
How CRISPR Immunity Works
spacer aquisition
“adaptation”
expression of crRNAs
interference via effector complex
Cas0
crRNA
tracr RNA
Protospacer Adjacent Motif (PAM)
[any nucleotide]GG next to the spacer sequence
must be present right next to the target DNA site for the Cas9 to bind and cut, acting as a critical "self/non-self" marker and limiting where editing can occur
not found in the CRISPR DNA array
simple and common elsewhere
Key Innovation of CRISPR
substitution of chimeric (combination of different) gRNA in place of natural crRNA and tracrRRNA
Genomic Editing with CRISPR
sgRNA designed to target a specific sequence in genome
Cas9 makes double stranded cut in genome
cellular DNA repair mechanism engaged: 2 possibilities
broken ends can be rejoined without any template (NHEJ)
broken ends can be rejoined using a template (HDR)
NHEJ
non homologous end joining is the most common type of repair to double strand break in DNA
no template used, which results in INDEL mutations
once mutation does occur, the resulting frameshifts lead to non-functional alleles gene silencing
knockout
HDR
homology directed repair is another way to repair double-strand breaks in DNA
uses same repair enzymes as in crossing over or recombination
can use a homologous chromosome (sister chromatids) as a template
in CRISPR experiments, can inject donor DNA at the same time as Cas9-CRISPR to stimulate HDR
INDEL
insertion and deletion mutations
CRISPR Advantages
relatively cheap and easy
can design single-guide RNA to target almost any sequence desired
relatively specific
indels created by non-homologous end-joining can create gene knockouts to determine gene function/phenotype
can be introduced to intact, living cells
can introduce Cas9 with donor DNA to stimulate HDR
CRISPR Challenges
Off-target effects: unspecific cleavage
Mosaicism
Non Specific Cleavage
challenge of CRISPR due to off-target effects
a modified Cas9 structure has been created to use a longer target sequence, but slower acting
can be hard to control whether NHEJ or HDR is used.
germline cells have enhanced HDR, adding donor template DNA may help
Mosaicism
challenge of CRISPR
not all cells are edited, so they get the mosaic effect
delivery of Cas9 is not 100% for all cells, and is a challenge for multicellular organisms
various approaches for delivery: transfection, microinjection, electroportation
embryo injections at single-cell stage
Uses of CRISPR
basic research
editing genomes to meet human desires
CRISPR Use: Basic Research
create gene knockouts
disrupt genes to determine unknown gene functions
sometimes, knocking out a gene results in a desirable phenotype
CRISPR Use: Hacking
editing, or hacking, genomes to meet human needs and desires
reversing mutations that cause genetic fisorders
donor organs from animals
improved farm animals
domestication of new plants for agriculture
de-extinction of extinct species
gene drives to eliminate insect-spread disease
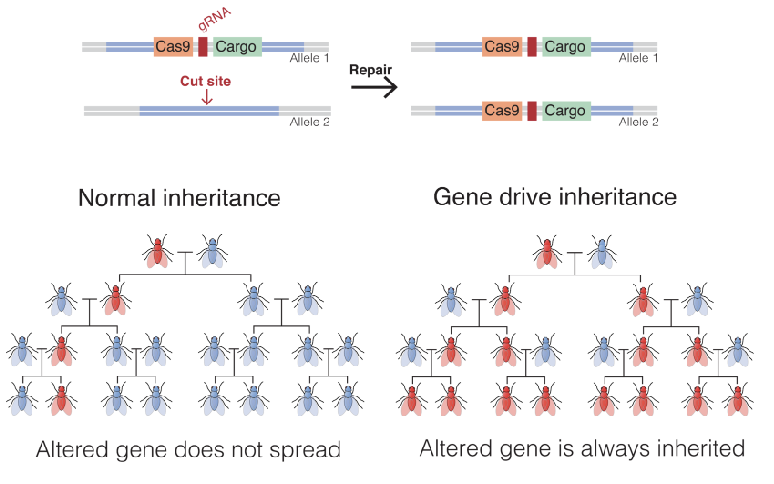
Gene Drives
a DNA construct, once introduced as one copy, copies itself to homologous chromosomes via HDR.
after sexual reproduction, heterozygous offspring are converted to individuals homozygous for gene drive construct
in this manner, the payload gene can be spread rapidly through the population, because of non Mendelian inheritance
could be used to insert gene for resistance to malaria, or a gene that reduces fertility of mosquito
Reasons for Cloning DNA in Living Cells
to make more DNA with high fidelity for further study or manipulation
to produce substances of scientific or commercial value from genes
to modify the genomes of plants or animals to introduce new, desired traits
Cloning Vectors
a DNA molecule, like a plasmid or virus, that carries a foreign DNA segment (gene of interest) into a host cell (like bacteria) to be copied (cloned) and amplified
common:
plasmid, specifically pUC 19
Plasmid Vector
Plasmids are commonly used as cloning vectors, as they are stable, self-replicating molecules which contain circular DNA
contains the:
origin of replication
selectable markers to identify cells that have taken up the plasmid
unique restriction enzyme cleavage sites
pUC 19 Plasmid
pUC 19 are typical bacterial vectors, as they contain a portion of the lacZ+ gene, with a restriction site linker that contains numerous unique restriction enzyme cut sites
unique sites= sites found nowhere else on the plasmid
Inserting Foreign DNA Sequence into a Plasmid
cut the foreign DNA with a restriction enzyme
cut plasmid with the same restriction enzyme
mix cut foreign DNA and cut plasmid DNA
use DNA ligase to seal sugar phosphate bonds
Competent Bacteria
bacterial cells that have been treated or are naturally predisposed to take up foreign DNA from their environment
E. coli made receptive to transformation by chemical or electrical treatment
lacZ- which lack the portion of the lacZ gene that is present in the plasmid
ligated plasmids containing DNA inserts are used to transform competent cells
Transformed Bacteria
bacterial cells that have taken up foreign DNA, usually a plasmid, through a process called transformation, making them acquire new genetic traits like antibiotic resistance
transformed competent cells plated out on agar media
bacteria with no plasmid do not grow
there is no antibiotic resistance
bacteria with non-recombinant plasmid produce B-galactosidase, resulting in blue colonies
bacteria with recombinant plasmids do not produce B-gala, resulting in white colonies
Bacterial Vectors Making Gene Products
includes operon and regulatory sequences to allow expression of genes in bacteria
good for production of many enzymes, especially those that originate from bacteria
Taq DNA polymerase
commercially available restriction enzymes
not good for gene products thats require post-transcriptional modification, as occurs with many eukaryotic proteins
Rhizobium radiobacter
naturally transforms the DNA of higher plants
can be co-opted to introduce new genes to plants
genes such as Bt
Bacillus thuringiensis
produces a protein, Bt toxin, which is lethal to many insects, but non toxic to humans and other animals
Bt gene has been transferred to many plans using R. radiobacter
Molecular Toolbox
Restriction endonucleases(REs) repurposed as DNA scissors.
Ligases allow DNA molecules (from different sources) cut by REs to be recombined to produce novel recombinant DNA, including
plasmids & other vectors that can transform other organismsPCR is a convenient way to make modest amounts of a particular DNA sequencing, but when large amounts of DNA, or the products of genes (enzymes, proteins) are required, cloning is best.
also routinely used to check success of expirements
Gel electrophoresis used routinely in combination with RE cleavage or PCR to check the success of cloning/transformation experiments.