Microbial Ecology Quiz 1
1/42
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
43 Terms
What does the Arrhenius equation tell us?
reactions get faster at higher temperatures!
there is an upper limit to this, but equation does not include that
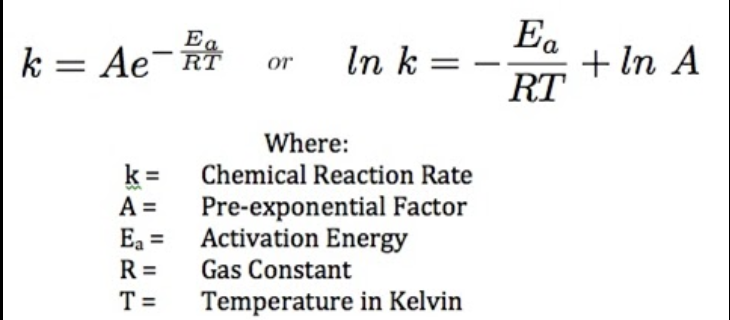
what is “activity”?
the “felt concentration” of a chemical in a mixture
not the same as concentration!! accounts only for available amount of molecules in solution
how do biomolecules react to extreme pH levels?
they will either lose H+ in very high pH (basic) conditions or gain H+ in very low pH (acidic)
this can change their charge and destabilize their structure
H-bonds are disrupted
which molecules are most likely to give up/accept H+ and why?
cysteine and histidine, because their pKa is near physiological pH (7)
what happens when there’s too much H+ outside the cell?
can break down the proton gradient that drives ATPase, and therefore prevents the production of ATP
what is pKa?
pKa = pH value at which a molecule’s dominant form shifts from one state of protonation to another
when at pKa, pH balances out more bc side-chains are absorbing protons rather than letting them float around surrounding environment
which types of molecules go through a cell membrane the easiest? which have the hardest time getting through?
small, nonpolar molecules go through easiest! (outside of cell membrane is made of nonpolar lipids)
ions have the hardest time getting through (strong charge, polar). need to get through membrane through some sort of pump
what are the 2 ways cells avoid damage due to pH?
removing protons from the cell (mitigation)
adapting to deal with it (adaptation)
how to cells remove protons to prevent pH damage?
enzyme-catalyzed reactions consume protons
reactions produce metabolites that soak up protons
ATPase can use ATP to pump protons out
how do cells adapt to avoid damage from extreme pH?
can change the membrane composition by adding my cyclopropane fatty acids
these lower proton permeability through the cell membrane
Chaperone proteins can re-fold proteins
ex. Hde activates in low pH and can refold partially unfolded proteins
proteins can perform DNA repairs
how might ATPase adapt to high pH conditions?
the c-ring of ATPase forms more subunits, allows protons to be pumped out when the proton gradient is weaker
what variables does water activity depend on?
molality of water in solution
activity coefficient of water

what might make water unavailable in an environment?
**anything that binds water lowers water activity!!
can be bound to salt crystals or other surfaces, which makes it unavailable to microbes
can also be bound between mineral layers
true or false: water activity changes depending on temperature
TRUE! 😄
increased temperature generally means increased activity
at what activity coefficient do most microbes inhabit?
ranges from 1.0-0.8 water active environments
0.6 is the lowest we know of, this is an extreme example though
what are 2 ways cells deal with water activity?
try to keep salt inside the cell (ex. a lot of KCl, Na+)
this is similar to how they deal with acidity! cell wants to be in equilibrium with salt content of surroundings
this requires significant changes on an evolutionary time scale
the “compatible solute” strategy (much more common)
cell produces small solutes that decrease water activity to match activity of surroundings
what are 2 examples of physical environments with high salt concentration?
deep hypersaline anoxic basins
salty “lakes” at bottom of water sources; water is much denser bc it’s very salty
salt pools in sf bay area (:D)
each pool has a different salt composition, different microbes are present in each one

what is redox potential (Eh)?
the tendency of a solution or environment to gain or lose electrons
positive Eh = potential to gain e-
negative Eh = potential to lose e-
what does a redox tower show us?
helps figure out what the cell is using to acquire e- and what it’s using to get rid of them
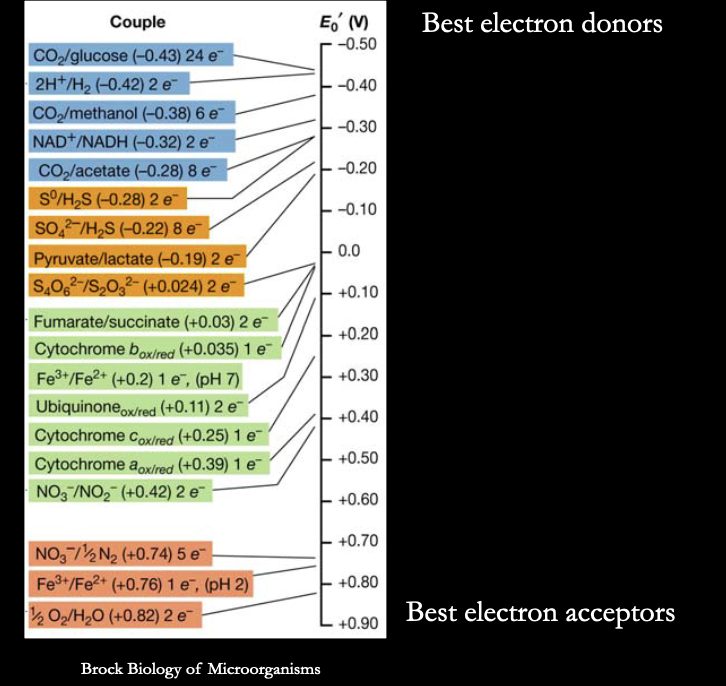
how do you read a redox tower?
e- falls from top of tower to the bottom (like potential energy! you get the most energy from falling the farthest)
each line is a redox couple (left is oxidized, right is reduced)
tells you what each molecule will turn into if you give it more e-
best e- donors are at the top, best e- acceptors are at bottom
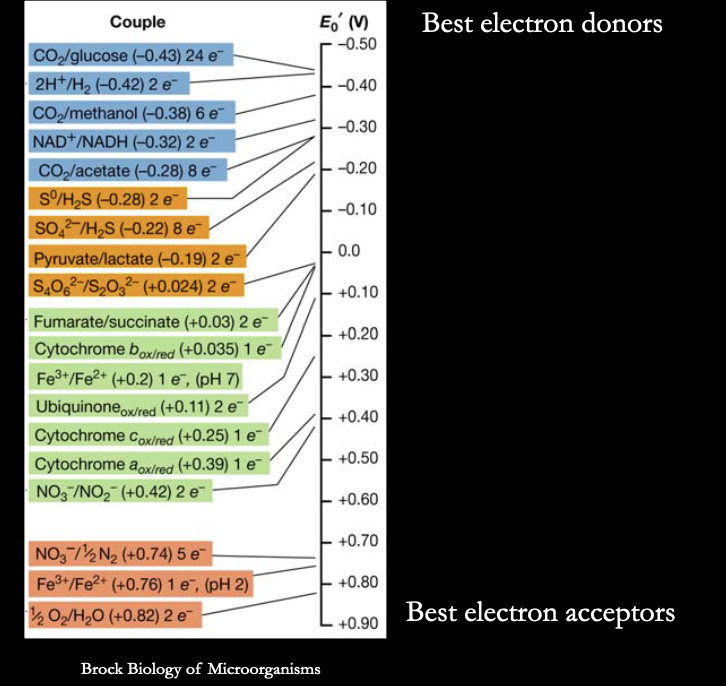
what does it mean for something to be reduced vs. oxidized?
reduced = has a lot of e-. and therefore can be a donor (can give them away!)
oxidized = has less e-, and therefore can be an acceptor (big and greedy)
reduced form of more negative potential couple donates e- to oxidized form of more positive potential couple
oxidation rules (OX #). take a deep breath it’ll be fine 😄
pure elements, O2 and H2 = O
H = +1
O = -2
K, Na, etc = +1
Cl, F, etc = -1
Na, Mg, etc = +2
reminder to do practice redox problems. god knows you need to <3
do it babes pls. instructions on lecture 4 slides
what distinguishes microbes from eukaryotes?
metabolic diversity
animals use glucose and O2 to create ATP, microbes can use a wide variety of pathways
carbon-fixation pathways (conversion of atmospheric Co2 → sugar to use for photosynthesis)
there’s 7 total, and 6 are only found in microbes

what does the arrow in a reaction pathway signify?
an enzyme catalyzing the reaction
specifically how enzymes + cofactors have multiple conformations to further a reaction
what is the Rate Law and what does it tell us?
Rate = k[A]^m [B]^n
describes how the speed of a chemical reaction depends on the concentration of its reactants
rate = speed of reaction (rate of substrate uptake or product formation)
k = rate constant, which is specific to temp and pH conditions (reflects the “efficiency” of a microbe’s enzymes
[A], [B] = molar concentrations of reactions (concentration of substrates like glucose, oxygen, nitrogen)
m, n = reaction orders which are determined experimentally (indicates sensitivity of microbe to changes in substrate availability)
![<p>Rate = k[A]^m [B]^n </p><ul><li><p><strong>describes how the speed of a chemical reaction depends on the concentration of its reactants</strong> </p></li><li><p>rate = speed of reaction (rate of substrate uptake or product formation)</p></li><li><p>k = rate constant, which is specific to temp and pH conditions (reflects the “efficiency” of a microbe’s enzymes</p></li><li><p>[A], [B] = molar concentrations of reactions (concentration of substrates like glucose, oxygen, nitrogen)</p></li><li><p>m, n = reaction orders which are determined experimentally (indicates sensitivity of microbe to changes in substrate availability)</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/82acb240-92f0-4c03-8e89-dc41c7fca719.png)
what does “0 order reaction” refer to?
the rate is independent of concentration of reactants
aka the reaction will happen at a constant speed regardless of how much reactant is present
this is most common when the available sites for a reaction to take place are limiting
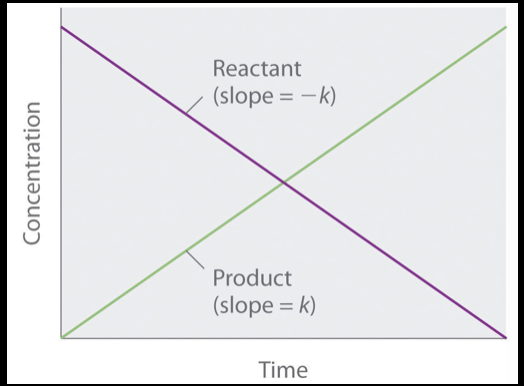
what does “1st order reaction” refer to?
the reaction rate is proportional to the concentration of one of the reactants
this is most common with a single limiting reactant, but there’s an abundance of reaction sites
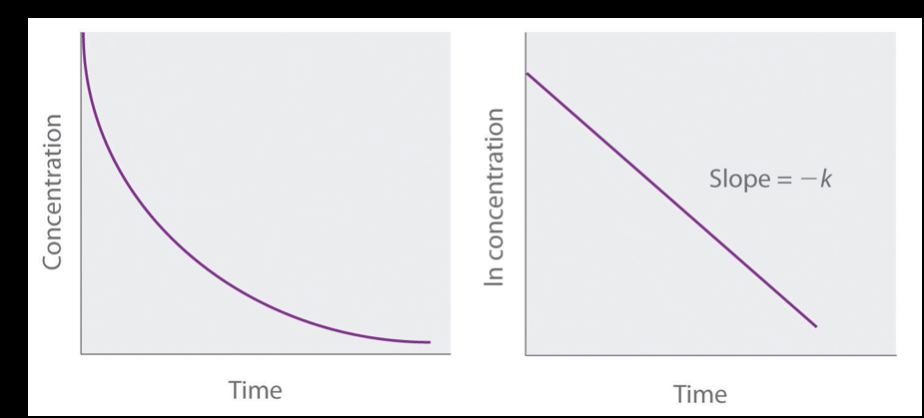
what does “2nd order reaction” refer to?
the reaction rate is proportional to the concentration² of one of the reactants OR the product of the concentrations of 2 reactants
in this case, if you double the concentration of reactants, the reaction speed will be 4x faster (2² = 4)!
this is most common when monomers become dimers
most of the redox metabolic reactions are a 2nd order reaction
how do enzymes speed up reactions?
holding substrates such that bonds are closer to the transition state
lowering entropy by bringing the reactants together
What is Vmax?
the rate of reaction when all enzyme active sites are full (maximum rate of reaction)
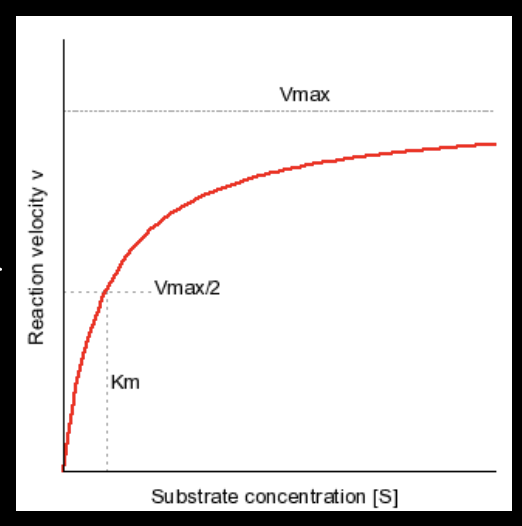
What is Km?
substrate concentration at which the reaction rate is exactly half of Vmax
demonstrates the “affinity” of an enzyme for it’s substrate
“affinity” refers to the strength of bond between enzyme and substrate
a smaller Km = higher affinity
bc you don’t need a high concentration of substrate to get the enzyme to work at half it’s max speed
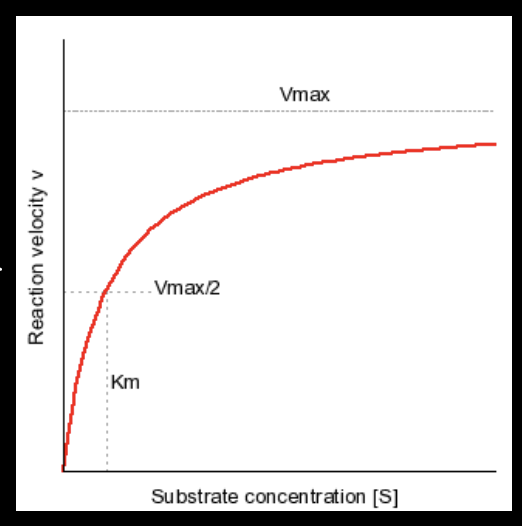
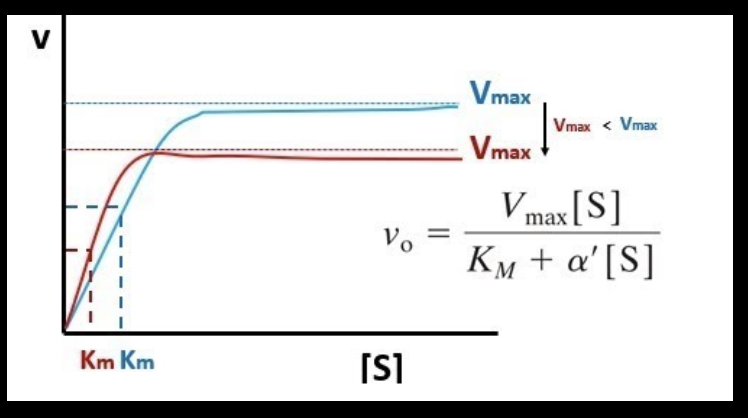
with low substrate concentration, would you expect to see more P2 or P3?
Km of the red line is smaller, meaning there’s a higher affinity! more P2 is expected
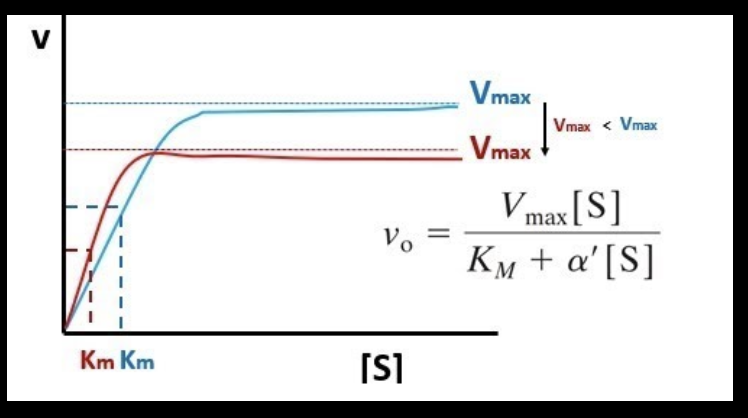
with high substrate concentration, would you expect to see more P2 (red line) or P3 (blue line)?
the blue line has a higher Vmax, so you would expect to see more P3
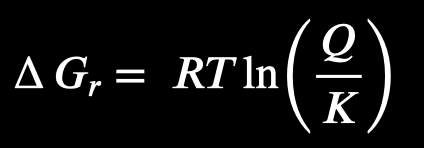
in the Gibbs energy equation, what do Q and K mean?
K = equilibrium constant (destination)
a fixed value for a specific reaction at a specific temperature
tells us the ratio of products to reactants when the reaction is at equilibrium
Q = reaction quotient (current location)
used concentration of products and reactants at any given moment
what can Gibbs energy tell us about a reaction?
Gibbs energy call tell us if a reaction is exothermic (can happen spontaneously) or endothermic (needs energy to happen)
-G = spontaneous, but doesn’t mean the reaction will definitely happen
relationship between Q and K tells you what direction the reaction needs to go to reach equilibrium
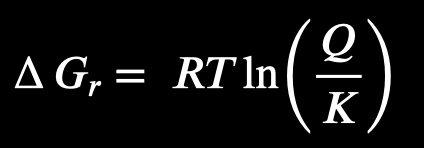
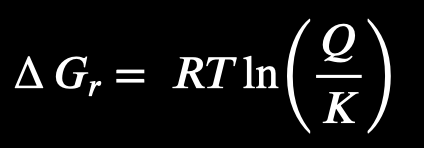
How can you predict if a reaction will happen based on the Gibbs energy equation?
Q<K:
the reaction is spontaneous, and will move forward to create more products
Q=K
the reaction is at equilibrium, and no net change in gibbs free energy is happening
Q>K
the reaction is not spontaneous (needs energy), reaction will move backward to create more reactants
what is maintenance energy?
the energy a cell consumes just to stay alive
for a cell to grow, the rate of energy production has to exceed the maintenance energy
“no-growth” condition occurs when energy source is equal to the maintenance energy
which type of amino acids is life on Earth made of?
left-handed amino acids
these can switch over time (this switch is called racemization)
racemization kinetics is the speed at which this switch happens!
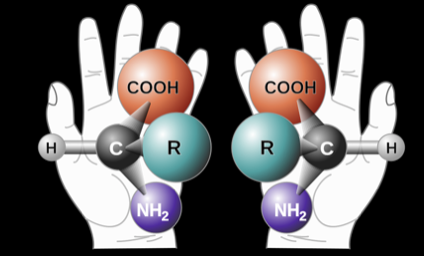
when do amino acids undergo racemization?
when an organism dies and it’s proteins are no longer maintained
over long periods of time left-handed amino acids
lower than expected racemization is an indication of microorganisms continuously degrade dead biomass to synthesize new left-handed amino acids
what are different interpretations of the term “growth”?
making new biomass
making more biomass
making more cells
what are different interpretations of the term “activity”?
metabolic activity
making new biomolecules
what can rRNA presence tell us about cell growth?
presence of rRNA indicates proteins being built, and therefore shows that cells are growing rather than just surviving
high RNA:DNA ratio shows high cell activity: abundance