Genetics Exam 2
1/72
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
73 Terms
What are the roles of genetic material?
Information: must contain info to make organism
Transmission: can be passed to offspring
Replication: must be copied to be passed to offspring
Variation: capable of change ot account for phenotypic variation
Frederick Griffith's Experiments with Streptococcus pneumoniae
Concluded that something from the dead type S (virulent) bacteria was transforming the type R bacteria into type S in mice where both were present, called this the transforming principle

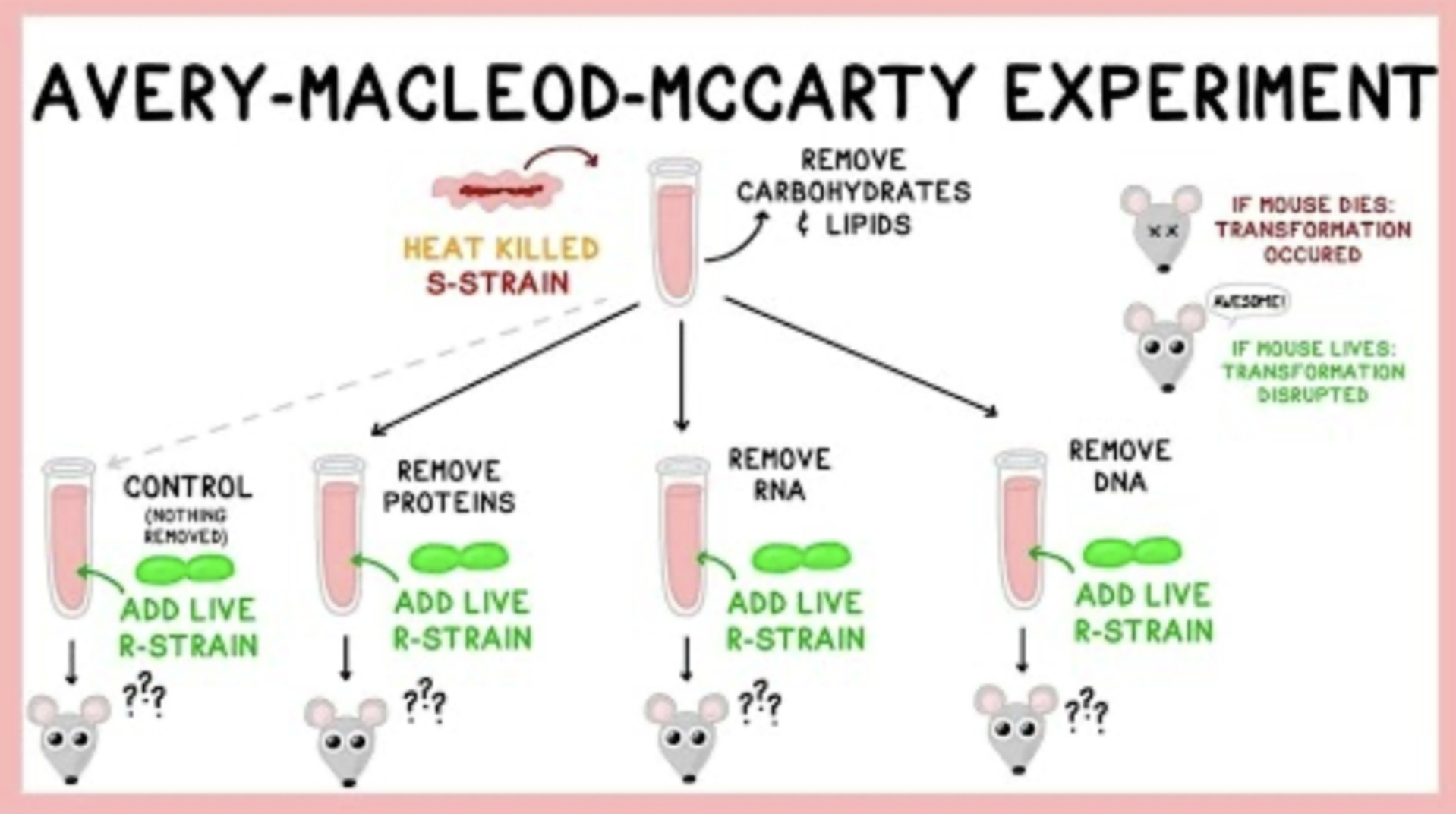
How did the experiments of Avery, MacLeod, and McCarty follow up that of Griffith?
Before experiment, weren’t sure if proteins or nucleic acids were the transforming principle
They selectively destroyed different molecules and showed that only the destruction of DNA prevented the transformation of harmless bacteria into virulent ones; the researchers identified DNA as the "transforming principle”
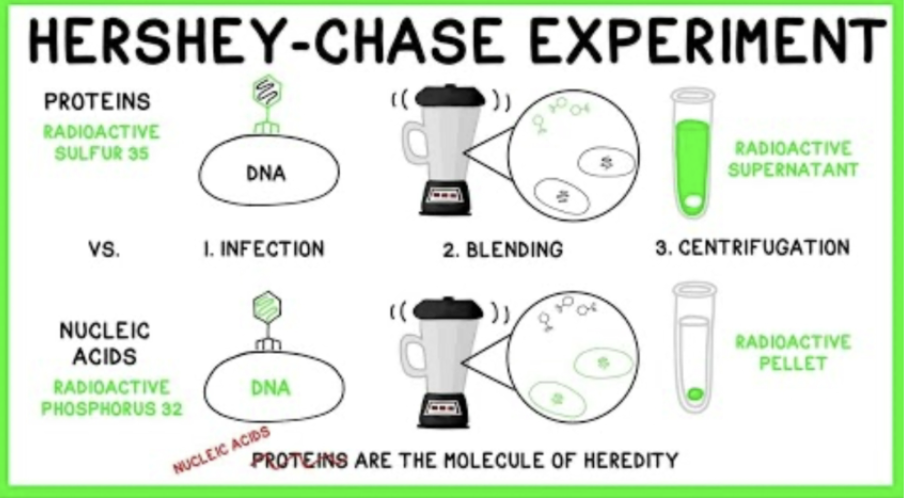
How did the experiment of Hershey and Chase follow up that of Griffith?
Hershey and Chase also later provided evidence that DNA is the genetic material of T2 phage
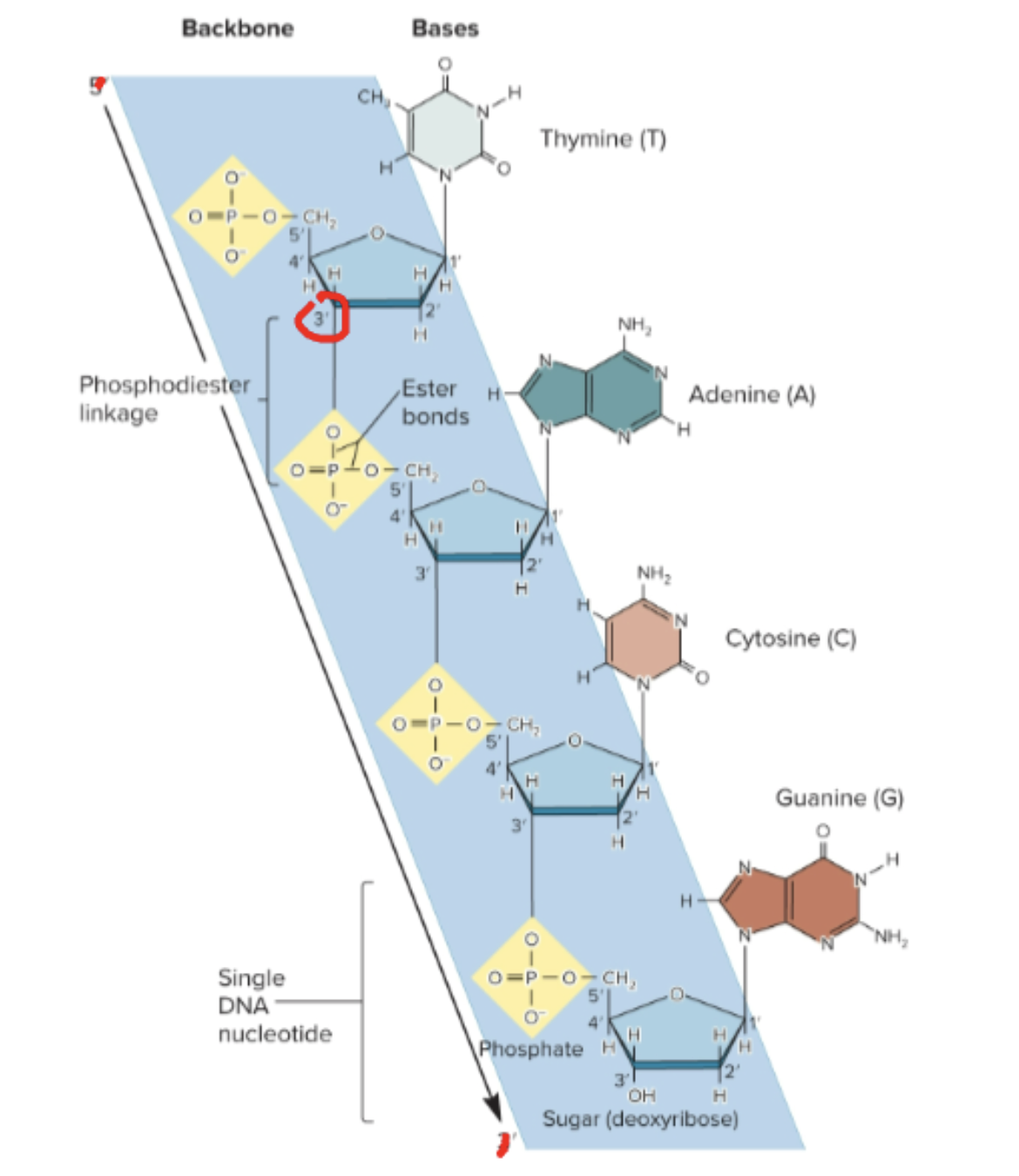
What are the three parts of a nucleotide?
A phosphate group, a pentose sugar (ribose in RNA and deoxyribose in DNA), and a nitrogenous base
What is the difference between purines and pyrimidines?
Purines have a double-ring base and include adenine and guanine
Pyrimidines have a single-ring base, and include thymine, cytosine, and uracil
What are the differences between DNA and RNA?
DNA is a double-helical structure stabilized by hydrogen bonding between complementary bases, while RNA is single-stranded and more flexible
RNA uses uracil instead of thymine
RNA uses ribose sugar, which has an extra oxygen atom in comparison to the deoxyribose sugar of DNA
RNAs are much shorter, being only several hundred to several thousand nucleotides in length
How are nucleotides linked together?
A phosphate connects the 5’ carbon of one nucleotide to the 3’ carbon of an adjacent one, called a phosphodiester linkage
What did Franklin’s experiments find?
The double helix structure of DNA
What are the general characteristics of the structure of DNA?
A is bonded to T by 2 hydrogen bonds, while C is bonded to G by 3 hydrogen bonds
2 asymmetrical grooves on the outside of the helix, called major and minor grooves, proteins can bind within these grooves
Can form secondary structures:
B DNA: right-handed helix (standard)
Z DNA: left-handed, backbone follows a zig zag pattern
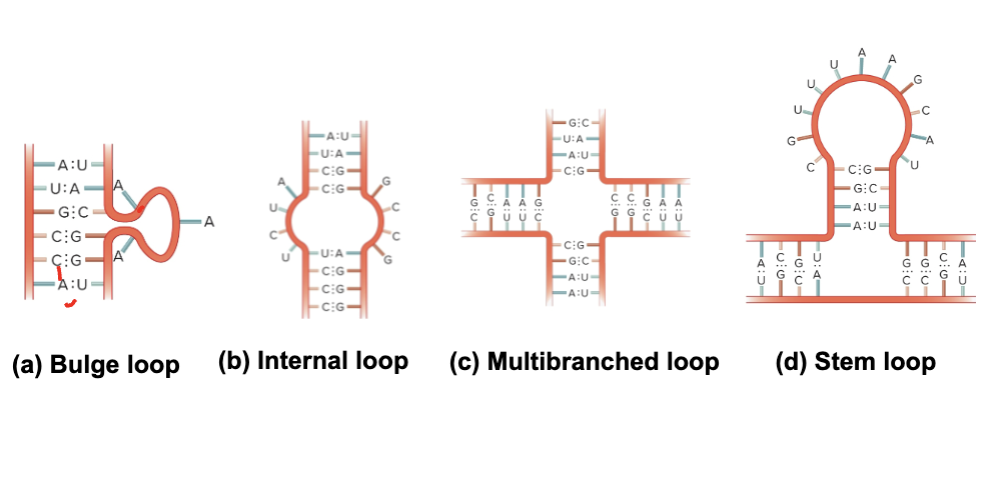
What are the secondary structures of RNA?
Bulge loop
Internal loop
Multibranched loop
Stem loop
Characteristics of prokaryotic chromosomes
Entire genome is one singular chromosome
Circular molecule that is a few million bps in length
Mostly protein-coding genes, with nontranscribed DNA called intergenic regions
Single type of chromosome, but may be present in multiple copies
How are bacterial chromosomes compacted?
Use DNA-binding proteins called nucleoid-associated proteins (NAPs) to form:
loop domains (microdomains)
adjacent microdomains organized into macrodomains
also play role gene regulation
How does the structure of archaeal chromosomes depend on the DNA-binding proteins they express?
Some archaeal species produce bacterial-like NAPs
Others produce eukaryotic-like histone proteins
Besides NAPs, what is another way that prokaryotic chromosomes become compact?
Supercoiling (formation of additional coils due to twisting forces)
Negative supercoil: to the right (loosening, increases transcription)
Positive supercoil: to the left (tightening, decreases transcription)
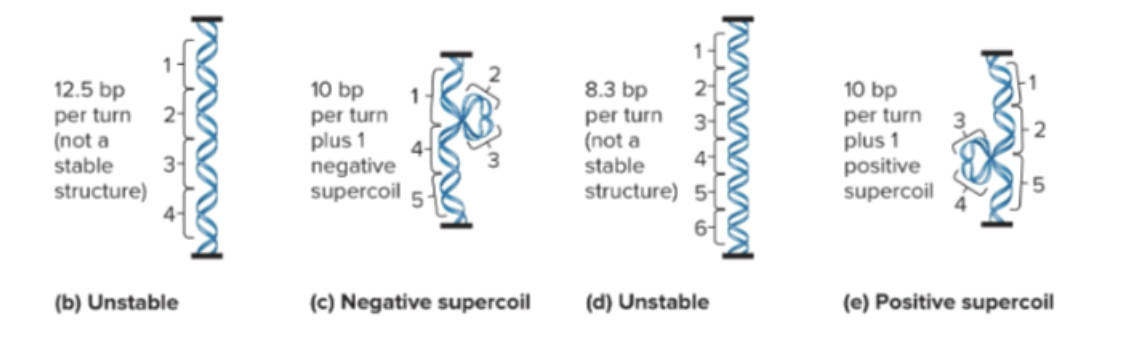
What enzymes control supercoiling in bacterial chromosomes?
DNA gyrase (DNA topoisomerase II): introduces negative supercoils using ATP, can relax positive supercoils, and can untangle intertwined DNA molecules
DNA topoisomerase I: relaxes negative supercoils
Why are genes longer in complex eukaryotes in comparison to simpler eukaryotes?
Complex eukaryotes have many introns (noncoding intervening sequences
What are the three types of DNA sequences required for chromosomal replication and segregation?
Origins of replication
Centromeres
Play role in segregation
Telomeres
important in replication and for stability
Where are repetitive sequences most commonly found?
Near centromeric and telomeric regions, but may also be interspersed throughout the chromosome
What is responsible for the variation in eukaryotic genome size?
Accumulations of repetitive DNA sequences
What are the three main types of repetitive sequences in eukaryotes?
Unique or non-repetitive: found once or a few times, includes protein-coding genes and intergenic regions
Moderately repetitive sequences: found a few hundred to several thousand times, includes genes for rRNA and histones, sequences that regulate expression and translation, and transposable elements
Highly repetitive sequences: found tens of thousands to millions of times, each copy is relatively short, some sequences are interspersed throughout the genome, others clustered together in tandem arrays
What is transposition?
The integration of small segments of DNA into a new location in the genome
The small, mobile, DNA segments are called transposable elements (TEs)
What are the two types of transposition pathways?
IR = Inverted repeat
The main difference between LTR retrotransposons and non-LTR retrotransposons is that:
Reverse transcriptase and integrase used for LTR
Non-LTR moves by a target site primed reverse transcription
What are the outcomes of transposition/why does it exist?
Possible just because they can, they can proliferate because they don’t harm (selfish DNA theory)
Or possibly that they offer an advantage, increasing variation and leading to evolution of new genes
Most outcomes are harmful
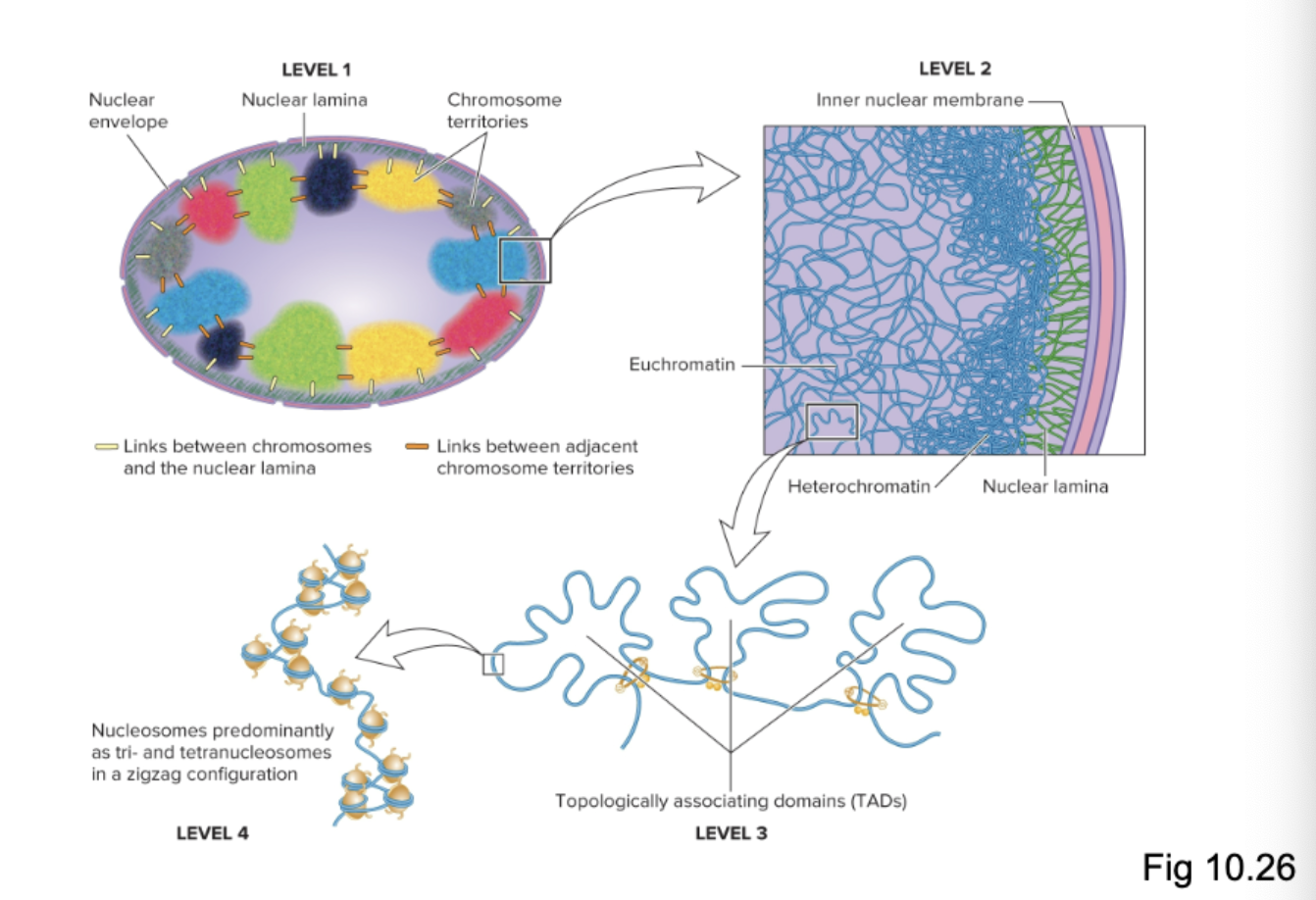
What are the four levels of organization of chromosomes in a non-dividing eukaryotic cell?
How can transposition inactivate a gene?
Transposition of a TE into the middle of a gene, making it nonfunctional (like the color producing gene in corn)
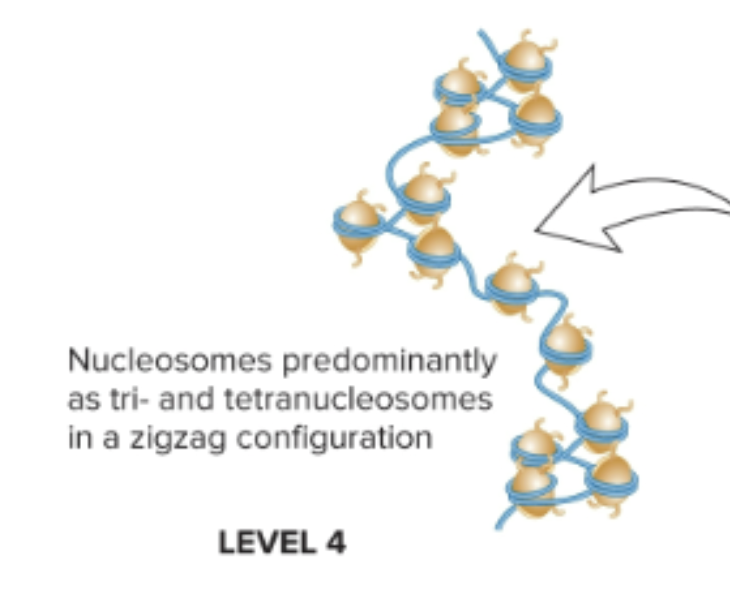
What is the smallest level (level 4) of chromosome organization in non-dividing eukaryotic cells?
Nucleosomes predominantly as tri and tetranucleosomes in a zigzag configuration
Nucleosome composed of a double-stranded segment of DNA wrapped around an octamer of histone proteins (H2A, H2B, H3, H4, and H1 aka linker histone)
What is the second smallest level (level 3) of chromosome organization in non-dividing eukaryotic cells?
Topologically associating domains (TADs)
Loops are formed by 2 proteins:
SMC proteins: form a dimer that can wrap itself around two DNA segments and promote the formation of a loop
CCCTC binding factor (CTCFs): two of them bind to dna and bind to each other to stabilize loop
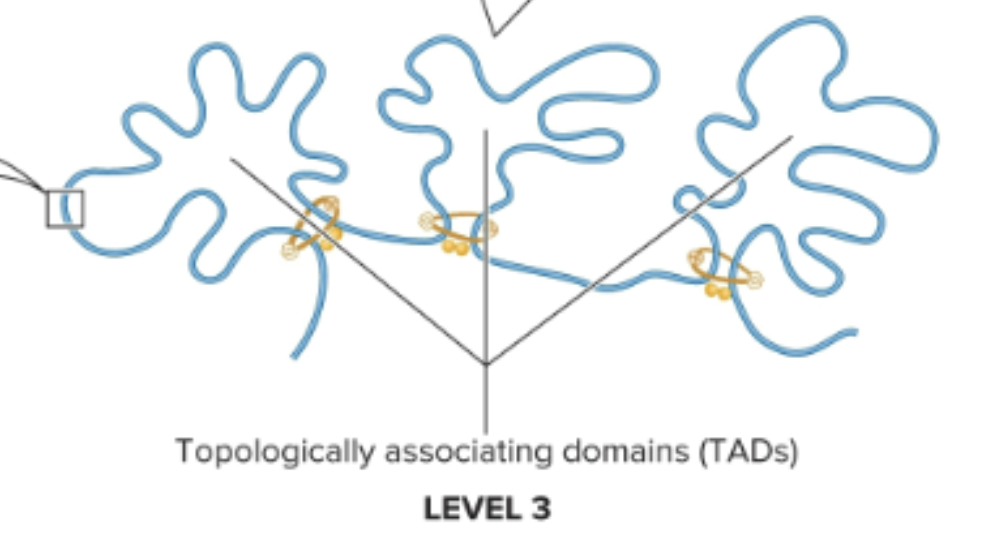
What is the second-largest level (level 2) of chromosome organization in non-dividing eukaryotic cells?
Heterochromatin (compact chromatin), euchromatin (less condensed)
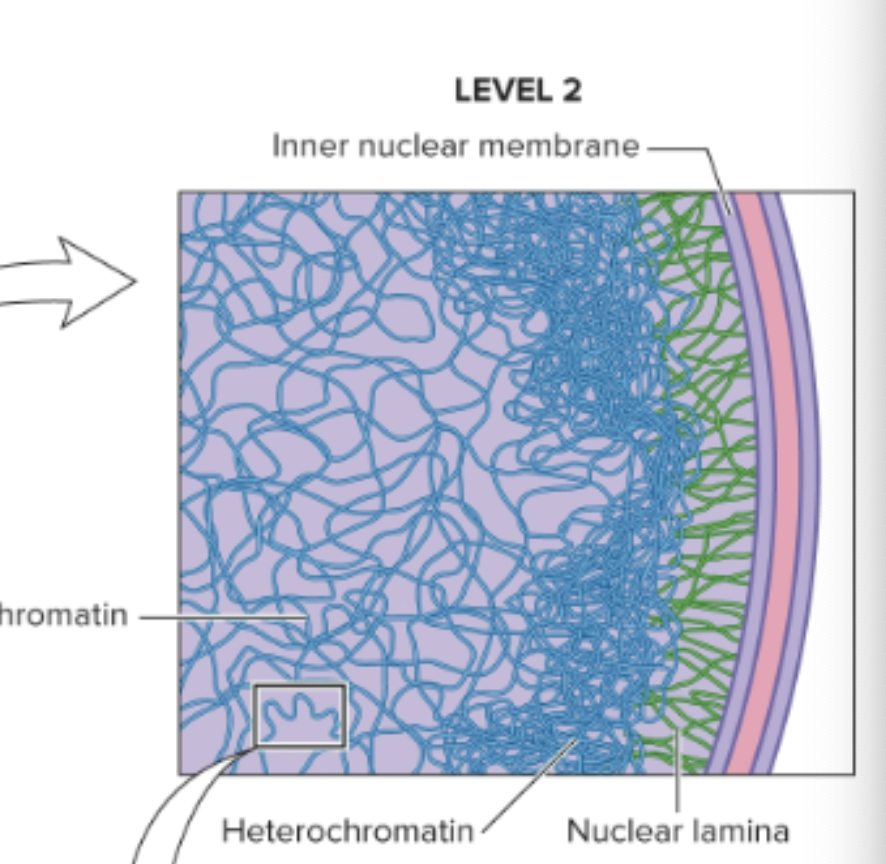
What is the largest level (level 1) of chromosome organization in non-dividing eukaryotic cells?
Chromosome territories
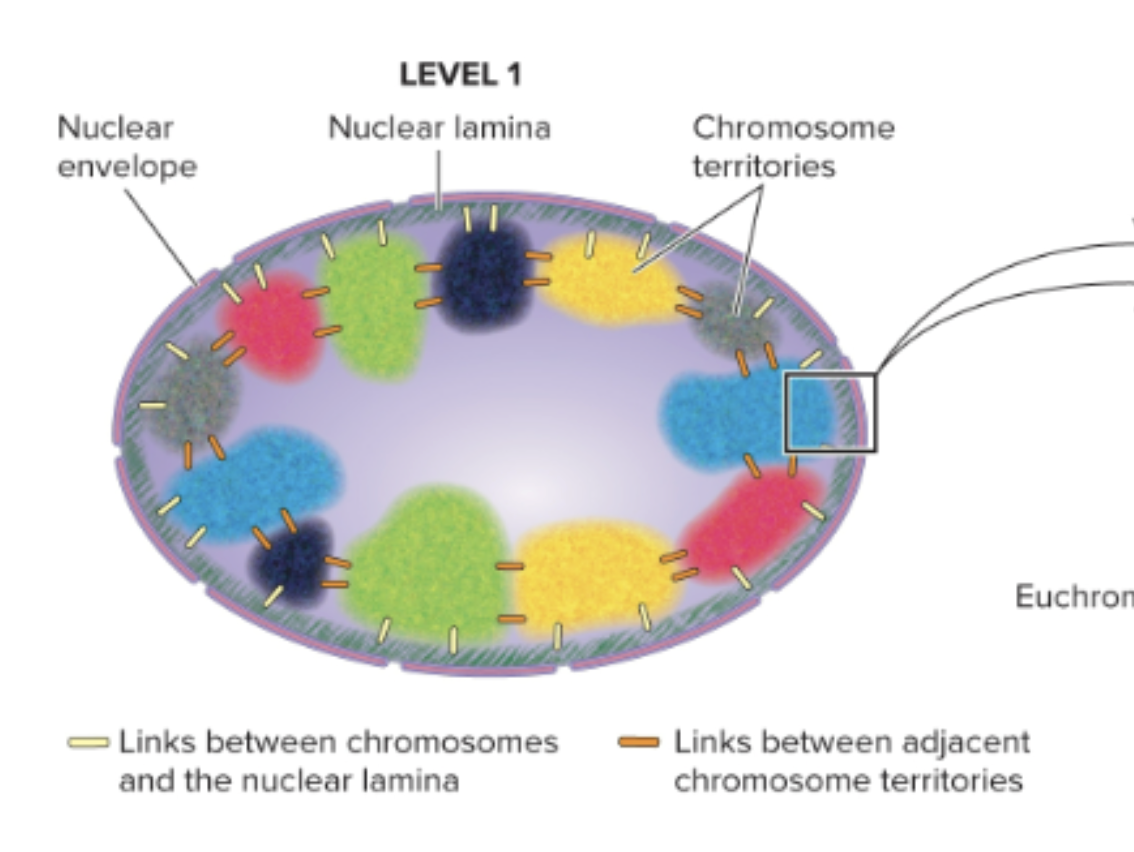
What are the 2 types of heterochromatin?
Constitutive: always heterochromic, and heterochromic at the same location in all cell types
Facultative: can switch between heterochromatin and euchromatin, and varies at a single location on chromosome depending on cell type
What two protein complexes help form and organize metaphase (compact) chromosomes?
Condensin (condensin I and II): plays a role in chromosome condensation
Cohesin: plays a critical role in sister chromatid alignment
Both contain a category of proteins called SMC proteins, use ATP to catalyze changes in chromosome structure
Three proposed models of DNA replication
Conservative: both parental strands stay together
Semiconservative: one parental and one daughter strand (correct)
Dispersive: parental and daughter segments interspersed
How was it determined that DNA replication follows the semiconservative model?
Meselson and Stahl used light and heavy nitrogen
Parental strands are heavy
Step 1 of bacterial DNA replication: Origin of replication
The origin of rep. in E. coli is called oriC
3 types of DNA sequences found at origin of rep. are functionally important
dnaA boxes: sites from the binding of dnaA protein, dnaA proteins also bind to each other, causing the DNA strand to bend and break
AT-rich regions: where DNA breaks (weaker bonds)
GATC methylation sites: sites that help to regulate DNA replication
DNA adenine methyltransferase (dam) methylates the A on both strands. Immediately after replication, the daughter strand is not methylated, takes a while, replication cannot occur yet
Step 2 of bacterial DNA replication: Synthesis of new DNA strands
DNA helicase breaks H-bonds between base pairs, which generate positive supercoiling ahead of fork
DNA gyrase travels in front of helicase to relax this supercoiling
Single-strand binding proteins prevent DNA from re-binding
Primase synthesizes RNA primers
DNA polymerase III catalyzes the attachment of nucleotides to make a new strand in the 5 to 3 direction aka it travels on template strand in 3 to 5 (alpha subunit does this)
DNA polymerase I removes the primers and replaces with DNA
DNA ligase catalyzes the formation of a covalent bond to connect the backbones
Once fork reaches tus protein bound to a ter sequence (T1 or T2), replication ends
DNA ligase covalenty links the 2 daughter strands, and DNA gyrase separates the 2 intertwined molecules (catenanes)
Primosome vs replisome
Primosome contains the primase and helicase
Replisome contains the primosome and the 2 DNA polymerases
Where does DNA pol catalyze a covalent?
Between the innermost phosphate group of the incoming deoxyribonucleoside triphosphate and the 3’-OH of the sugar of the previous deoxynucleotide
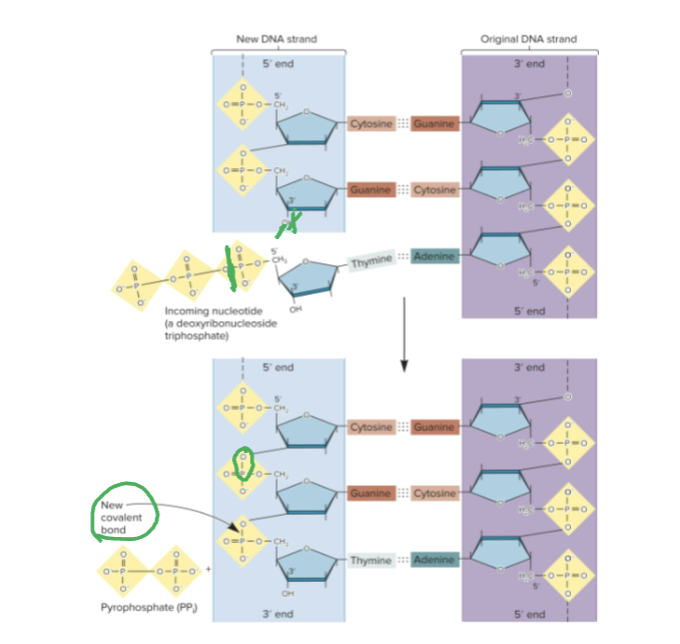
Alpha vs beta subunit of DNA pol III
Alpha subunit catalyzes the attachment of nucleotides
Beta subunit forms a dimer in the shape of a ring around template DNA, termed the clamp protein
Fidelity mechanisms of DNA replication
Complementary pairs have much higher stability than mismatches
Helix distortion caused by mispairing prevents an incorrect nucleotide from fitting properly in the active site
DNA pol uses exonuclease activity to digest the newly made strand until the mismatched nucleotide is removed (proofreading)
How does eukaryotic DNA replication differ from prokaryotic?
Multiple origins of replication
Origins of replication are more complex
Are more dynamic
Contain G-quadruplexes
Promoters and CpG islands are frequently found in the nucleosome-free region
Begins with the assembly of the prereplication complex (preRC), and includes the origin recognition complex (ORC) which is a 6 subunit complex that acts as the 1st initiator of eukaryotic DNA replication
Flap endonuclease removes RNA primers
Telomerase adds DNA sequences to telomeres since the end of the strand cannot be replicated since there is no room for primer, which causes shortening of the telomere
Multiple types of DNA polymerase that synthesize DNA
What are the 3 main classes of origins of replication in eukaryotes?
Constitutive: always used
Flexible: used in a random manner, most common type
Dormant: used during cell differentiation or only at a specific stage of development
Types of DNA polymerase in eukayotes
Alpha: works immediately after primer
Epsilon: used on leading strand
Delta: used on lagging strand
What are the two types of regulatory transcription factors (RTFs)?
Repressors: bind to DNA and inhibit transcription
Activators: bind and increase transcription
Negative vs positive control
Negative: regulation by repressors
Positive: by activators
What are small effector molecules and what are the types?
They affect the rate of transcription but do not bind to DNA directly; instead bind to TFs
Inducers: increase transc.
corepressors and inhibitors: reduce transc.
Corepressors bind to repressors and cause them to bind to DNA
Inhibitors bind to the activator and prevent it from binding to DNA
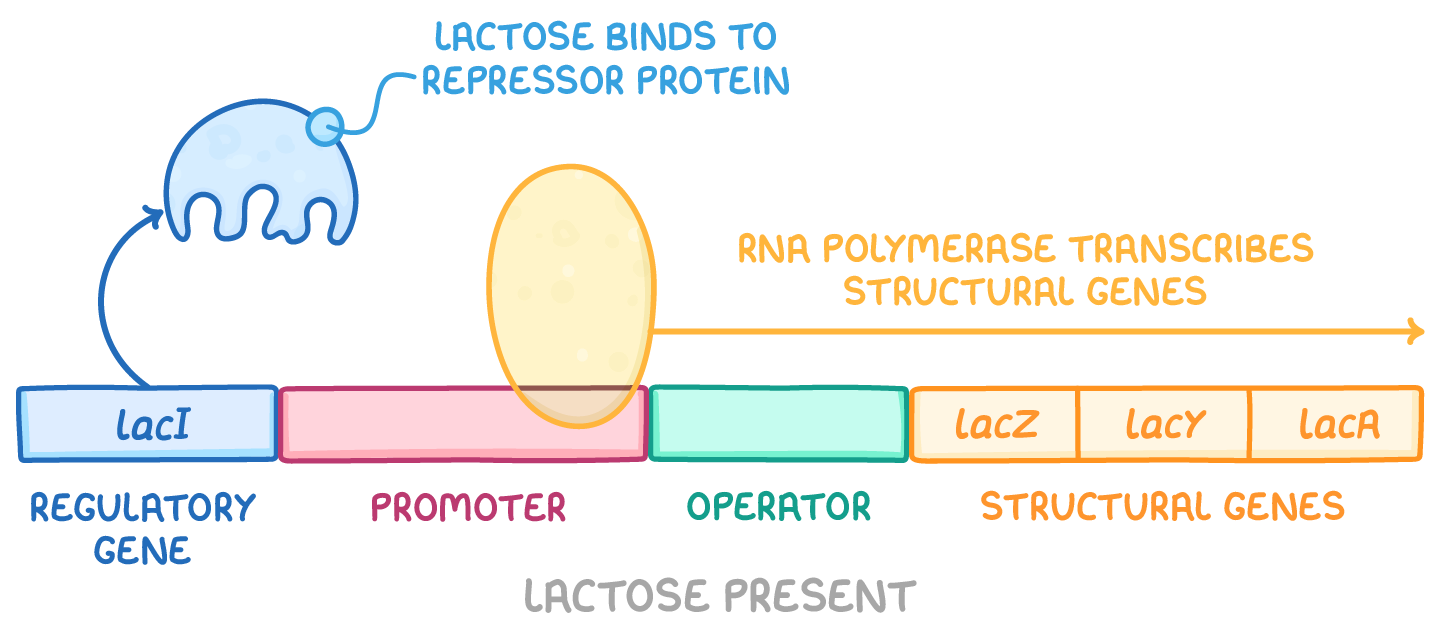
What is the lac operon and how does it work?
Regulatory elements:
Promoter: binding site for RNA polymerase to start transcription
Operator: binding site for lac repressor protein
CAP site: binding site for catabolite activator protein (CAP) which helps enhance trasc. when glucose is low
Regulatory genes
lacl: encodes the lac repressor protein
Structural genes:
lacZ: encodes beta-galactosidase, which breaks down lactose
lacY: encodes lactose permase
lac A: encodes thiogalactoside transacetylase, which removed toxic byproducts
When lactose is present:
Some lactose is converted to allolactose, which binds to lac repressor and prevents it from binding to DNA, increasing trasc.
When no lactose is present:
No allolactose is present so the repressor stays bound
When glucose is not present:
cAMP is present in high levels, binds to the activator (CAP) and increases transc.
trans vs cis effect in regulation
Trans-effect: genetic regulation that can occur even though DNA segments are not physically adjacent
Cis-effect: a DNA sequence that must be adjacent to the genes it regulates
What are riboswitches? What is an example?
RNA that exists in 2 different secondary conformations: one is active, the other inhibits gene expression
Conversion between the two is due to the binding of a small molecules
Example: regulation of transcription of thi operon in B. subitlis
Active form of thiamin is thiamin pyrophosphate (TPP)
Regulation of TPP biosynthetic enzymes occurs through a riboswitch that controls translation
Low TPP: transc. is completed, shine-dalgano sequence binds
High TPP transc. reduced, it cannot bind
What are enhancers in eukaryotic regulation?
DNA region that contains 1+ regulatory elements
Regulatory TFs recognize cis-regulatory elements within enhancers
Regulatory elements are bidirectional/orientation independent
In what ways can the function of RTFs be modulated in eukaryotes?
Binding of a small effector molecule
Protein-protein interactions
Covalent modification
What is chromatin remodeling? What are histone variants? What are histone modifications?
Histones can change locations to regulate transc. by changing access to genes, some transcriptional activators do this
A few histones have accumulated mutations, making variants, some promote open chromatin conformation, some promote closed
Some enzymes modify the amino-terminal tails of histones, which can:
influence the interaction between the DNA and histone proteins (histone code), providing a binding site for proteins to alter chromatin structure
What is DNA methylation and how is it used in regulation?
Covalent attachment of methyl groups carried out by DNA methyltransferase
Usually inhibits transcription
In vertebrates and plants, many genes contain CpG islands near promoters that can be methylated, which may influence binding of TFs or chrom. remodeling
Where are nucleosome-free regions (NFRs) most commonly found?
Beginning and end of eukaryotic genes
What are the steps of transcriptional activation in eukaryotes?
Note: occurs at enhancers or near core promoter
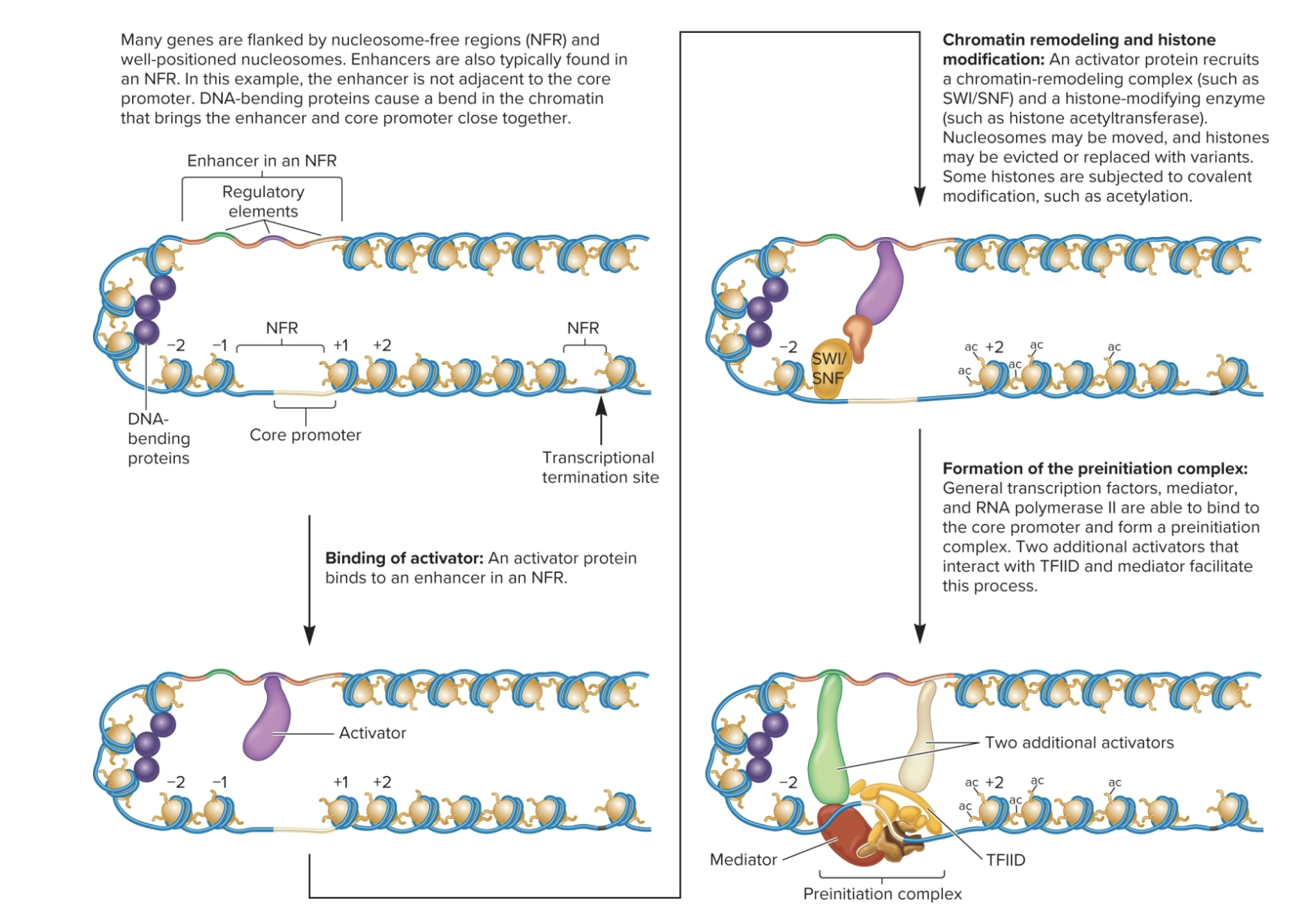
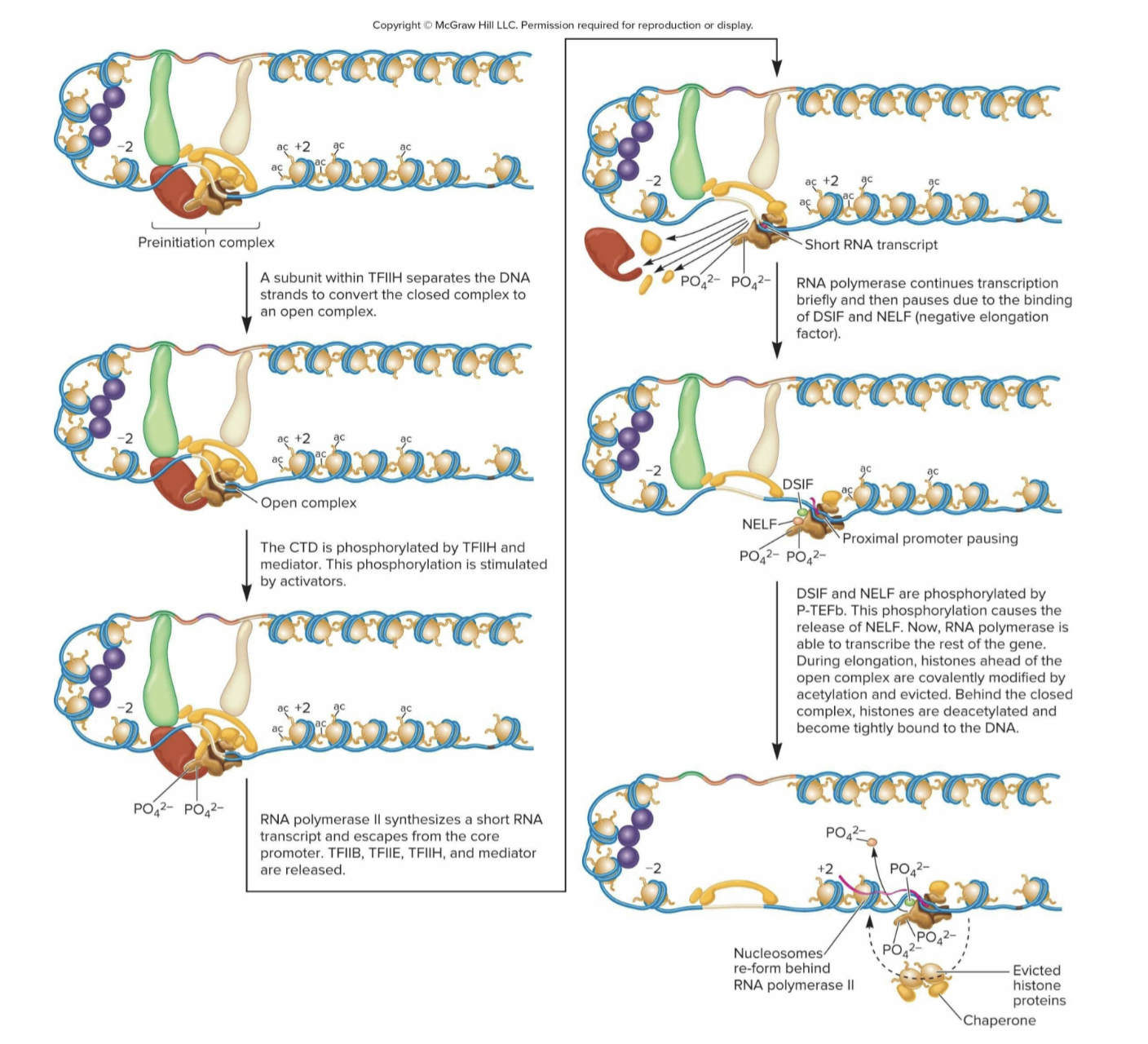
What are the steps of transcriptional elongation in eukaryotes?
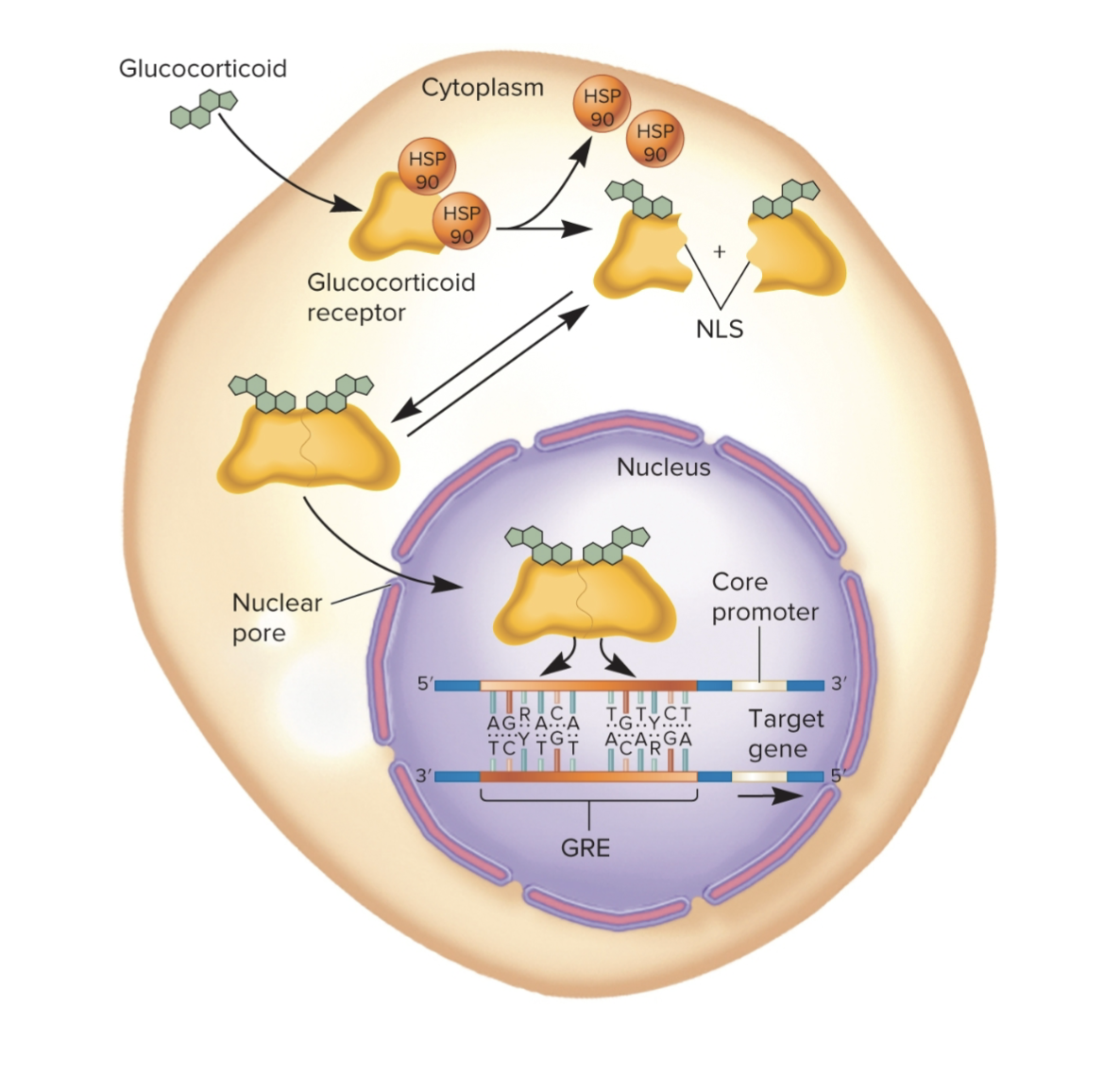
Glucocorticoid example of preinitiation complex
Note: presence of glucocorticoid increases transc.
CREB example of preinitiation complex
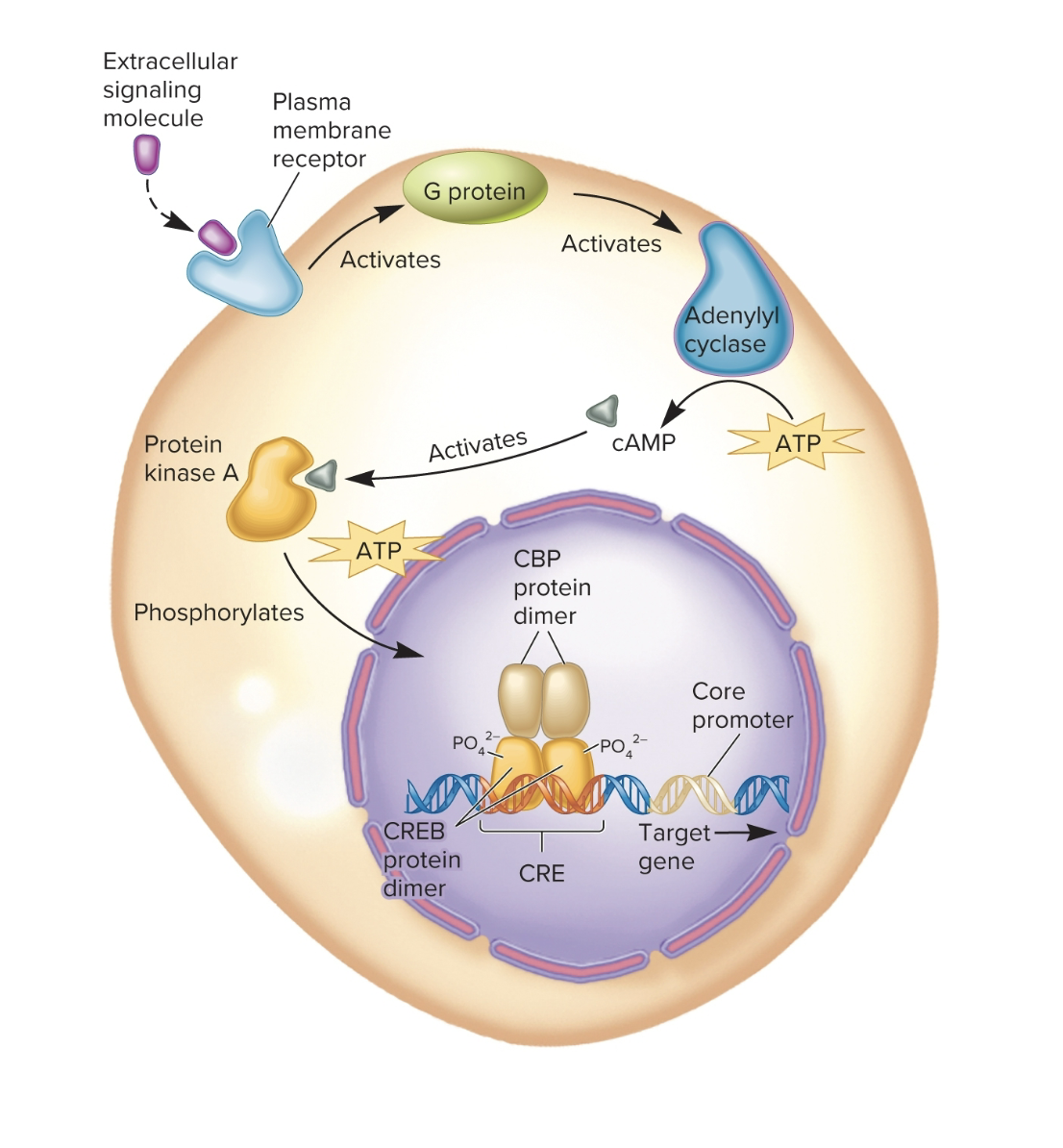
Besides transcription factors, what other way can genes or chromosomes be targeted for epigenetic regulation?
non-coding RNAs (ncRNAs)
Epigenetic regulation may occur as programmed developmental change or environmental agents
cis vs trans epigenetic changes
Cis: are maintained at a specific site, for example, may affect only one copy of a gene but not the other copy
Trans: maintained by diffusible factors, such as TFs, affects both copies of a gene
Facultative heterochromatin
Can switch between heterochromatin and euchromatin, and varies at a single location on chromosome depending on cell type
At multiple sites between the centromere and telomere, LINE-type repeats, methylation at CpG islands in gene regulatory regions, histone modifications (H3K9Me3, which causes repression)
What are post-translational modifications to amino-terminal tails?
result in changes in chromatin structure, such as heterochromatin formation
specific proteins bind to particular PTMs in nucleosomes via protein domains called reader domains
What are the higher-order structural features of heterochromatin?
Higher-order structure is the tiered changes in chromatin structure that allow for it to become even more compact. The three higher-order structures that occur are:
Having a closer, more stable contact of nucleosomes with each other via HP1 proteins (forms a dimer between modified H3 histones to shorten the distance between nucleosomes)
Forms closer loop domains
Binds to the nuclear lamina
Liquid-Liquid Phase Separation (sometimes)
What are the 3 phases of heterochromatic formation?
Nucleation: short chromosomal site bound by chromatin-modifying enzymes and chromatin remodeling complexes
Spreading: adjacent euchromatin is turned into heterochromatin
Barrier: spreading stops when it reaches a barrier
What are some methods to maintain epigenetic marks?
Hemimethylated DNA becomes fully methylated via maintenance methylation
Histones recruit remodeling protein complexes to daughter chromatids
Higher-order structure favors reformation of heterochromatin
What are the three ways that changes that occur during development are maintained by epigenetic variation?
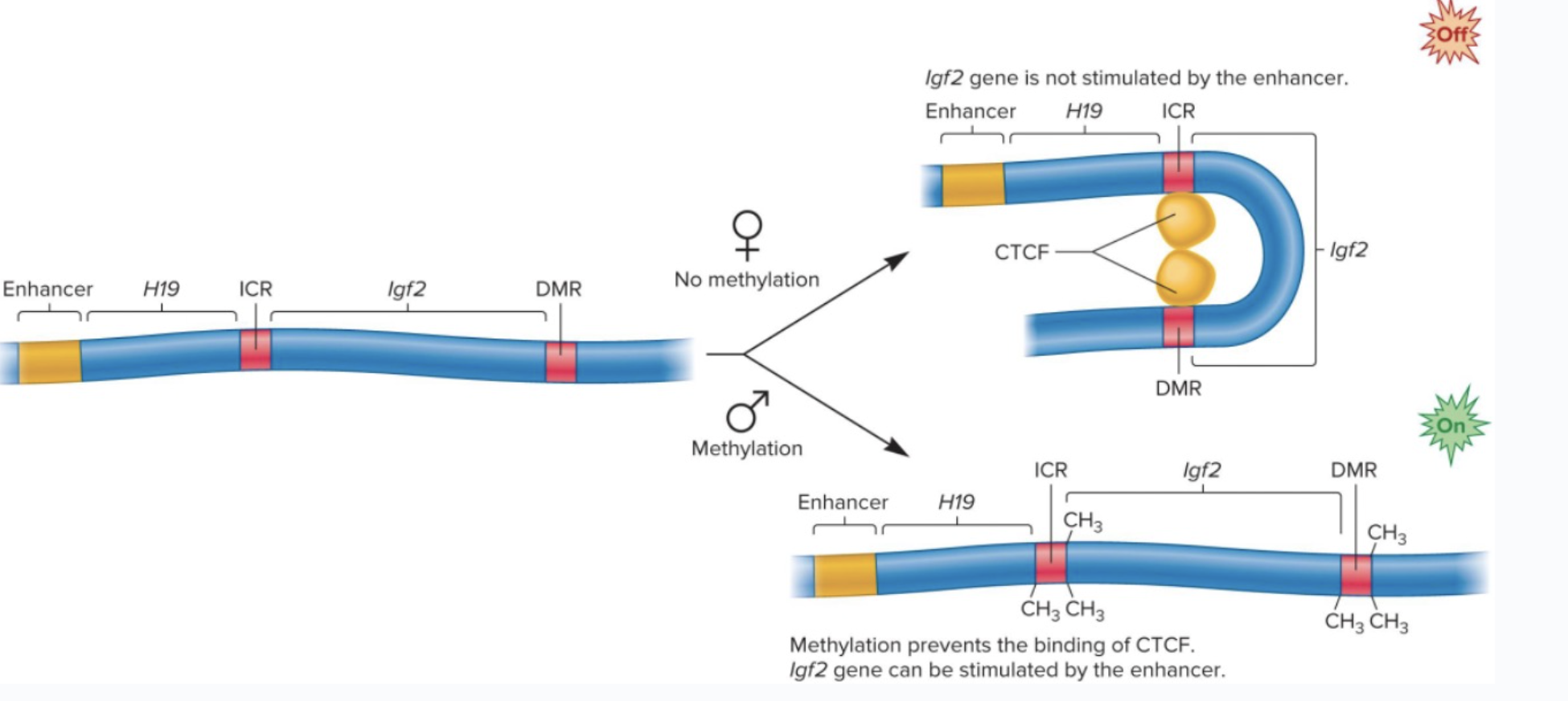
Genomic imprinting
offspring expresses copy from only one parent
X-chromosome inactivation
in a random manner, Tsix transc. continues. on one X chromosome, and Xist continues on other which causes Barr body formation
Epigenetic changes occur during embryonic development that are remembered during subsequent cell divisions
What are pioneer factors?
a category of TFs that can recognize and bind to DNA sequences exposed on the surface of a nucleosome, can recruit others to create a nucleosome-free region, involved in methods of activation and silencing
What two types of competing protein complexes are key regulators of epigenetic changes during development that produce specific cell types and tissues?
Trithorax group (TrxG): involved with activation
Polycomb group (PcG): involved with repression
PRC1 and PRC2 (binding of PRC2 to a polycomb response element leads to trimethylation of lysine 27 on histone H3, PRC1 inhibits by chrom. compaction, covalent modification of histones, or direct interaction with a TF)
How is the agouti gene in mice an example of epigenetics and environmental agents?
Agouti gene promotes synthesis. of yellow fur pigment
When fed a diet that increases DNA methylation, they were less yellow
Also example of royal jelly in bees causing a decrease in methylation, leading to bees developing into queens
What are the function of non-coding RNAs?
Scaffold
Guide
alteration of protein function or stability
ribozyme (catalytic function)
blocker
decoy
What is HOTAIR?
Hox transcript antisense intergenic RNA
A PRC2 complex binds to 5’ end and LSD1 complex binds to 3’ end
HOTAIR binds to GA rich region next to a target gene
PRC2 trimethylates H3K27 and LSD1 demethylates H3K4, which may directly inhibit transcription, or they may lead to further changes in chromatin structure that inhibit transc.
How do ncRNAs affect translation? (NOT transcription)
RNA interference: a phenomenon in which double-stranded RNA (sense and antisense) causes silencing of mRNA, mediated by:
siRNAs: originate from exogenous sources (not normally made by cells), can enter from virus or experimentally, perfect match in bases to mRNA
miRNAs: endogenous, usually inhibit through partial complementarity
RISC: double-stranded RNA molecule produced from pre-miRNAs and pre-siRNAs by dicer, one strand will be degraded and strand not degraded is bound to proteins, this complex binds to mRNA to inhibit
What are the 3 stages of the CRISPR system in bacteria?
Adaptation: cas1 and cas2 protein complex cleaves bacteriophage DNA, the piece inserted into the CRISPR gene
Expression: exposure results in expression of the crispr, tracr, and cas 9 genes, create complex
Interference: guides to bacteriophage DNA and cleaves it causing it to inactivate