exam 3
1/195
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
196 Terms
What are polymorphisms in the human genome?
Polymorphisms are locations in the genome where different people have different base-pair sequences.
Do most polymorphisms change phenotype?
No — most polymorphisms do not affect phenotype; only some correspond to disease alleles.
Where in the genome are polymorphisms commonly located?
Many are found in introns, silent mutations, or non-coding regions.
Why are polymorphisms useful as identification markers?
They vary between individuals and PCR can detect specific alleles, making them strong ID markers.
What are the four major types of polymorphisms and which two are emphasized in class?
Types: SNPs, InDels, SSRs, CNVs. Focus: SNPs (most common; many disease alleles) and SSRs (used in DNA profiling).
Why are SNPs important in genetics and medicine?
They are the most common polymorphism and most disease alleles are SNPs; also used for genetic genealogy (e.g., 23&Me).
Why are SSRs useful in forensic science?
SSRs vary greatly in repeat number between individuals, making them ideal for solving crimes through DNA profiling.
What is an SNP?
A single nucleotide polymorphism is a polymorphism where individuals differ at a single base pair in the genome.
How often does a SNP locus occur?
About every 10 bp in the genome (mostly in genes).
Approximately how many SNP loci exist in humans?
About 300 million SNP loci.
How much SNP variation exists between two individuals?
They differ at about 1 in every 1000 bp (~3 million SNP differences).
What does it mean that most SNPs are biallelic?
Most SNPs have only 2 possible base variants out of A/T/C/G. This is because SNP starts with one mutation at a single nucleotide, and that mutation changes one base into another. Now the population has two versions at that position, the original base and the mutated base.
Where do SNP mutations come from?
Random DNA replication errors or exposure to mutagens.
In what genomic regions are SNP loci primarily found?
SNPs occur mainly in genes (especially introns and synonymous codons).
How much of the genome is intergenic “junk DNA”?
Roughly half of the genome consists of repeated elements between genes.
What types of repeats make up junk DNA?
SSRs (simple sequence repeats) and transposable elements
Which repetitive regions have functional roles?
Centromeres and telomeres.
How often does an SSR locus occur in the genome?
About every 30,000 bp.
Roughly how many SSR loci are in the genome?
About 100,000 SSR loci.
What percent of the genome consists of SSRs?
3%
What is the size of an SSR repeat unit?
2-10 bp.
What is the total length of an SSR locus?
4–200 bp.
What defines different alleles of an SSR locus?
The number of repeat units (copy number variation).
SNP vs. SSR location differences?
SNP variants: base changes inside genes (often introns).
SSR variants: repeat-number changes outside genes (junk DNA).
What causes SSR copy number to expand or shrink?
Slipped mispairing during DNA replication.
Does every person have SNP and SSR loci in the same genomic positions?
Yes — the loci are in the same places, but the alleles vary.
Do SNP and SSR variants always affect phenotype?
No. Their effect depends on whether the mutation occurs in a gene and where within that gene.
What is a nonanonymous mutation?
A mutation that affects gene function, usually because it alters coding sequences, promoters, splice sites, or causes harmful repeat expansions.
What SNP and SSR variants tend to affect gene function?
SNPs: promoter mutations, splice-junction mutations, codon changes
SSRs: repeat expansions inside genes (e.g., fragile X, Huntington)
What is an anonymous mutation?
A mutation that does not affect gene function, usually because it occurs in introns, synonymous codons, or outside genes.
What SNP and SSR variants typically do not affect gene function?
SNPs: synonymous codons, intronic SNPs
SSRs: repeat number changes outside genes
What is the relationship between variant frequency and functional effect?
Rare variants usually have functional effects (haven’t spread through population).
Common variants usually don’t affect function (neutral mutations).
Which of the following is an example of a SNP?
A. Individuals with Huntington disease have more trinucleotide repeats in the coding region of the HD gene than normal individuals.
B. A single base near the neurofibromatosis gene can be a G or a T but the phenotype is not affected.
C. The most common cystic fibrosis allele has a deletion of three base pairs in the coding region of the CFTR gene.
B. A single base near the neurofibromatosis gene can be a G or a T but the phenotype is not affected.
Before PCR, how were specific genomic fragments cloned?
By making a genomic library in bacteria, then screening colonies to find the one containing the desired DNA fragment.
Why was genomic library screening difficult?
Because researchers had to identify which bacterial colony contained the exact fragment they wanted.
What technology replaced bacterial cloning for copying DNA fragments?
PCR (polymerase chain reaction).
Who invented PCR and why was it revolutionary?
Kary Mullis, awarded the 1993 Nobel Prize. PCR allows rapid, in-vitro amplification of specific DNA sequences without using bacteria.
What does PCR accomplish?
PCR produces billions of copies of a specific DNA region, even if only one copy is present initially.
What sequences do you need to know in order to design PCR primers?
About 20 bp at each end of the region you want to amplify.
How far apart can PCR primers be for successful amplification?
Up to ~1000 bp apart.
How much template DNA is required for PCR?
DNA from one somatic cell (2 copies) or one gamete (1 copy) is sufficient.
How do we design primers now compared to before the Human Genome Project?
We now use the online human genome sequence to choose primers targeting any genomic region.
What materials are required to perform PCR?
Template DNA
Two primers (one for each strand)
dNTPs
Taq polymerase
Buffer
Thermal cycler
Why does PCR use Taq polymerase instead of human DNA polymerase?
Taq polymerase comes from Thermus aquaticus (hot springs) and is heat-stable, so it survives the ~95°C denaturation step.
What are the three main steps in a PCR cycle and their approximate temperatures?
Denaturation (~95°C) – DNA strands separate
Annealing (50–60°C) – primers bind to template
Extension (72°C) – Taq polymerase synthesizes new DNA
What is the purpose of PCR?
PCR makes billions of copies of a specific DNA region through repeated cycles of DNA synthesis.
How does the number of DNA molecules change during PCR?
DNA doubles each cycle (exponential amplification).
If a double-stranded DNA molecule is amplified by PCR, how many double-stranded molecules exist after four cycles?
A. 6
B. 8
C. 16
D. 32
E. 64
C. 16
Why can PCR amplify DNA from even a single cell?
Because PCR exponentially amplifies the target sequence, starting from even 1 DNA molecule.
How much sequence must be known to design primers for PCR?
About 20 nucleotides at each end of the target region.
What is the maximum distance between forward and reverse primers for efficient PCR?
Up to ~1000 base pairs.
Why is primer design rapid and easy today compared to before the Human Genome Project?
Because the entire human genome sequence is online, so any region can be easily targeted.
Why does PCR yield billions of copies?
Each cycle doubles the number of molecules, leading to exponential growth (e.g., 2 → 4 → 8 → 16 …).
What occurs during the denaturation step of PCR?
High heat breaks hydrogen bonds, causing double-stranded DNA to separate. This results in two single strands that can serve as templates for replication.
What happens during the annealing step of PCR?
Primers base-pair with complementary sequences flanking the target region.
What happens during the extension step of PCR?
Taq polymerase adds nucleotides to the 3’ end of each primer, synthesizing new DNA.
Why was PCR a major breakthrough in molecular biology?
It replaced bacterial cloning, made rapid DNA amplification possible, and transformed research, medicine, and forensics.
What is the rule for designing PCR primers?
Choose ~18–20 nt at each end of the target sequence, written 5'→3', with the reverse primer being the reverse complement of the bottom strand.
What is the key principle to remember when designing primers?
DNA polymerase extends from the 3′ end of primers, so primers must hybridize outward, flanking the region being amplified.
How does PCR help identify disease alleles?
PCR amplifies a specific exon or region so it can be sequenced to check for disease-causing mutations (SNPs or SSRs).
How does PCR allow genetic screening of IVF embryos?
At the 6–10 cell stage, one cell is removed, its DNA amplified by PCR, and disease alleles are detected. Embryos without disease alleles can then be implanted.
Why does IVF allow avoidance of inherited disorders?
PCR detects whether each embryo carries a disease allele; only wild-type embryos are implanted.
What is the genetic cause of sickle-cell disease?
A SNP in the β-globin gene (HBB) that changes GAG → GTG, replacing Glu with Val.
How does PCR help detect the sickle allele (Hb^S)?
PCR amplifies the HBB exon containing the SNP, and sequencing reveals whether the allele is A (normal) or T (sickle) at the SNP position.
How do homozygous vs. heterozygous SNPs appear in sequencing traces?
Homozygote: one clean peak (A or T).
Heterozygote: double peak at the SNP (both colors), because PCR products of both alleles are present.
Why do heterozygotes show mixed peaks at a SNP during sequencing?
Both alleles were amplified by PCR, producing two strands of the same length that differ at only one base → detector reads both nucleotides at that position.
Why can the same PCR primers amplify both alleles of a SNP?
Primers anneal outside the SNP and do not overlap the variant; both alleles share the same flanking sequence.
How does sequencing determine genotype at an SNP locus?
One peak → homozygous
Two peaks → heterozygous
Half of the PCR products end in one base, half in the other.
How do dideoxynucleotides reveal SNP alleles?
The chain terminates with a labeled ddNTP. If two ddNTPs appear at the same position → two alleles.
What distinguishes HBB^A from HBB^S in DNA sequence?
HBB^A: …C T C… (Glu)
HBB^S: …C A C… (Val)
One A↔T base change.
How can PCR detect SNPs from a single embryonic cell?
Because PCR amplifies the target region exponentially, producing billions of copies even from a single molecule.
What are the steps for detecting a SNP disease allele with PCR + sequencing?
Amplify region containing SNP.
Sequence PCR products.
Look at the SNP position:
One peak → homozygote
Two peaks → heterozygote
Interpret allele as disease or wild-type.
What genetic feature causes Huntington disease?
A trinucleotide SSR expansion (CAG repeats) in the Huntingtin (HTT) gene.
How many trinucleotide repeat disease genes exist?
17 known genes—including Huntington disease.
What does the repeating CAG codon encode in the Huntington protein?
Glutamine (Gln).
What is the normal length of the poly-glutamine tract in Huntington?
About 20–25 glutamines.
How does PCR detect Huntington disease alleles?
PCR amplifies the repeat region; product size reveals repeat length via gel electrophoresis—no sequencing needed.
What repeat lengths correspond to normal, late-onset, and early-onset HD?
Normal: ~20 repeats
Premutation: ~30 repeats
Late-onset disease: ~50 repeats
Early-onset, severe: ~100+ repeats
Why are PCR products longer in HD disease alleles?
Because disease alleles contain more CAG repeats, increasing the amplified fragment length.
What method detects SSR allele variation?
PCR using primers that flank the repeat + gel electrophoresis to measure product size.
What pattern appears on the gel for homozygous vs. heterozygous SSR genotypes?
Homozygous: one band (same size repeats).
Heterozygous: two bands (two different repeat sizes).
What determines how long the PCR product is for an SSR locus?
The number of repeats between flanking primers.
Why do SSR expansions indicate disease status?
Disease alleles have extra repeats, producing longer PCR fragments.
What does CODIS stand for and what does it do?
Combined DNA Index System — the FBI database using 20 SSR loci to identify individuals.
Why are SSRs ideal for forensic DNA profiling?
They are highly polymorphic, with ~10 alleles each, creating trillions of possible profiles—far exceeding the human population.
How many autosomal SSR markers are used in CODIS?
20 autosomal SSR loci, plus markers on X/Y for sex determination.
Why is it nearly impossible for two people to share the same DNA profile?
Because the probability of sharing the same alleles at 20 highly variable SSR loci is astronomically low.
What defines an individual’s forensic DNA fingerprint?
Their allele sizes (repeat numbers) at each of the 20 CODIS SSR loci.
What is a trinucleotide repeat disease, and how common are they?
A disease caused by expansion of a 3-bp SSR (simple sequence repeat) in a gene. There are 17 known trinucleotide repeat diseases (rare among 100,000 SSR loci).
Which SSR is expanded in the Huntington (HD) gene, and what amino acid does it encode?
CAG repeat; each CAG encodes glutamine (Gln).
How do homozygotes and heterozygotes appear on an SSR gel?
Homozygous: One band
Heterozygous: Two bands (each SSR allele yields a different-sized PCR product)
What is multiplex PCR?
A PCR method that amplifies multiple loci at once using several primer pairs in the same reaction.
Why does forensic DNA profiling use multiplex PCR?
Crime scene DNA may be scarce; multiplexing allows testing of many SSR loci simultaneously without needing lots of sample.
How do we distinguish different PCR products in multiplex PCR?
Each primer pair is labeled with a different fluorescent color; size differences separate alleles.
What does each band in a DNA fingerprint represent?
A person’s maternal or paternal SSR allele at that locus.
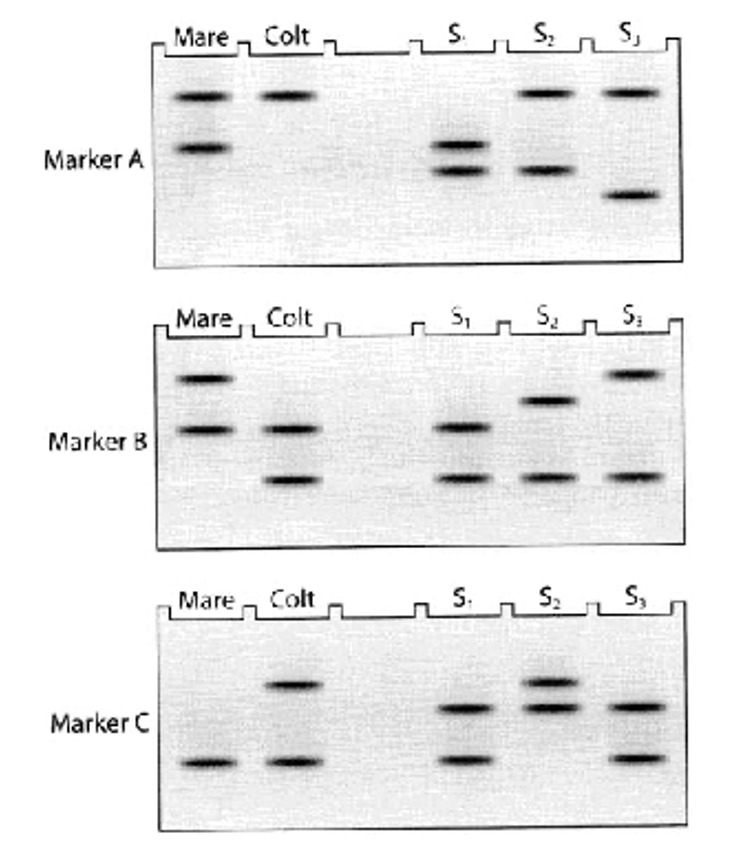
Which male stallion fathered the colt based on SSR markers?
Options:
A. Stallion 1
B. Stallion 2
C. Stallion 3
B — Stallion 2
Because the colt’s bands must match one allele from the mare and the other allele from the father.
Why can siblings have completely different SSR profiles?
Because of independent assortment — each child randomly inherits one of the two alleles from each parent at each SSR locus.
How did SNPs help solve the Golden State Killer case?
Police uploaded crime scene DNA to genealogy databases, matched distant relatives using shared SNP haplotype blocks, and built a family tree to identify the suspect.
What is a haplotype block?
A long stretch of linked SNP alleles inherited together due to low recombination (few crossover hotspots).
What does the length of a shared DNA segment between two people indicate?
Longer shared segments = closer genetic relationship.