M11L1 - Apoptosis
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
20 Terms
Why does regulated cell death occur?
Normal part of cell life cycle
It’s an eqm where you gain and lose as a constant process
Helps in preventing cancer i guess by not having too little cell loss
Apoptosis in embryo hand/feet development
Even in early development, apoptosis takes place
In week 6, skin forms webbing between the digits
By week 11, the webbing disappears due to apoptosis
Proof of Apoptosis in Embryo Webbing (Mouse Paw)
Mouse paw embryo stained with a dye to detect apoptosis
Shown as yellow dots seen mostly where the webbing is
As webbing disappears, the bright spots do too
This is done through TUNEL assay
TUNEL Assay in Apoptosis Detection
Takes advantage of the nicks present in apoptosis cell’s DNA
dUTP can be incorporated into the nicks by enzymes
Enzyme: Terminal Deoxynucleotidyl Transferase
Flourescently-labelled dUPT can specifically detect cells in apoptosis
Apoptosis in Frog Metamorphosis
Tadpoles undergo metamorphosis to become a frog
The tail disappears as the cells are induced to undergo apoptosis
This is stimulated by the increase of the thyroid hormone in blood
Apoptosis in Human Nervous System Development
Half of the cells originally produced are required in normal brain development
The other half undergo apoptosis
Cells that haven’t achieved synaptic connections
Cells with faulty connections
Cells not having made contact with a target cell
Matches the number of nerve cells with the number of target cells
Inappropriate Cell Death Diseases
Alzheimers: Neurons in hippocampus and cerebral cortex die
Huntingtons: Neurons in striatum die
Parkinsons: Dopamine neurons in substantia nigra die
Duchenne Muscular Dystrophy: Muscle cells die
What are the 2 ways in which cells die
Necrosis
Apoptosis
Necrosis
Cell death through damage to exterior
Cells swell and release contents to surrounding tissue
Can lead to infection
Apoptosis
Programmed cell death that is regulated
Cells suicide in response to stress/damage or as a part of normal development
The debris isn’t released to damage cells nearby
The debris is contained and recycled
Apoptotic Pathway
Cell Execution: Kill the cell
Engulfment: Get rid of the body
Clearance: Destroying the evidence
Ultrastructural Features of Apoptosis (7)
Chromatin compacts and condenses
Nuclear envelope breaks down
Nucleus contents are fragmented and the DNA / proteins are degraded
Cytoplasm undergoes condensation as cellular components aggregate
Mitochondria is permeabilized and released into the cytosol
Cell membrane moves and changes shape to create blebs (protrusions)
Cell fragments create compartments with debris which will be phagocytized and recycled
C. elegans Apoptosis Model
They’re studied very much in detail
947 somatic cells have been identified in the adult worm
The lineage of them all is traced to a single cell undergoing rounds of division
131 cells undergo apoptosis
Apoptosis Genes Identified by C. elegans model
Done by assay for identifying mutations in genes
These genes are called “cell death genes” (ceds)
Mutation in ced-1: Allows apoptosis but not the associated phagocytosis
Mutation in ced-3: No apoptosis observed
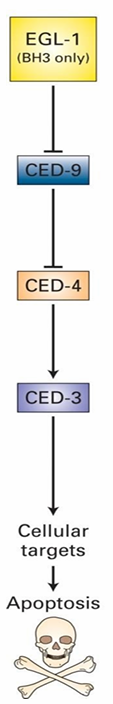
Four essential genes:
ced-3
ced-4
ced-9
egl-1
Mammalian Apoptotic Pathway
EGL-1 Homologs: Bid and Bim
CED-9 Homolog: Bcl-2
Bcl-2 controls Bak and Bax
CED-4/3 form a complex called the caspase holoenzyme
Protease targeting many different proteins for degradation
CED-3/4 mutations prevent death
ced-9 mutations make all cells die
Inhibits activation of caspase holoenzyme
Inhibits apoptosis in this was
EGL-1 signals apoptosis by inhibiting CED-9
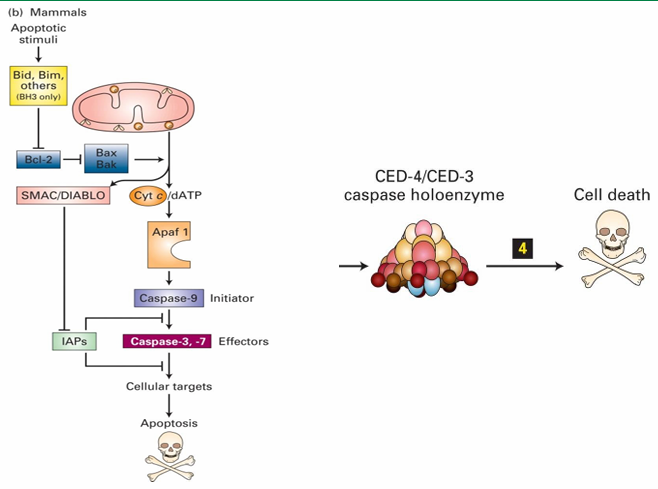
Caspase Holoenzyme
Apoptosome in mammalian cell
Contains direct homologues of C. elegans proteins
Apaf 1 = CED-4
Caspase-9 = CED-3
Protease activity of caspase holoenzyme leads to protein degradation and cell death
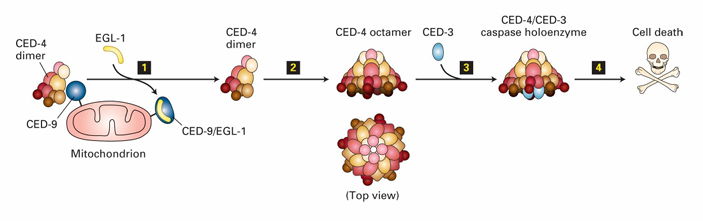
Activation of Caspase Holoenzyme (C. elegans)
In C. elegans
CED-9 inhibits apoptosis by binding to CED-4 dimers
Keeps them inactive
EGL-1 binding to CED-9 releases CED-4
CED-4 then join with CED-3 to form caspase holoenzyme
This leads to degradation of cytosolic and nuclear proteins
Mammalian CED-9 homologue
It’s Bcl-2
Normally anchored to outer membrane of mitochondria
Alters permeability of it
It maintains low permeability when present
When inactive, it forms pores associated with apoptosis
Bad: Apoptosis Signaling Pathway with Cytochrome C.
Mammalian cell apoptotic signal is called Bad
It’s inactive while phosphorylated and bound to 14-3-3
14-3-3 is a cytosolic adaptor protein
Signalling pathways allow dephosphorylation of Bad
It then releases from 14-3-3
It then binds to Bcl-2 on mitochondria
This activated Bcl-2 to allow for Bax to be activated
Bax aggregated into clusters in the membrane to make pores
Pores increase membrane permeability
Allows release of mitochondrial proteins into cytosol
This includes cytochrome C which is essential in forming mammalian apoptosome
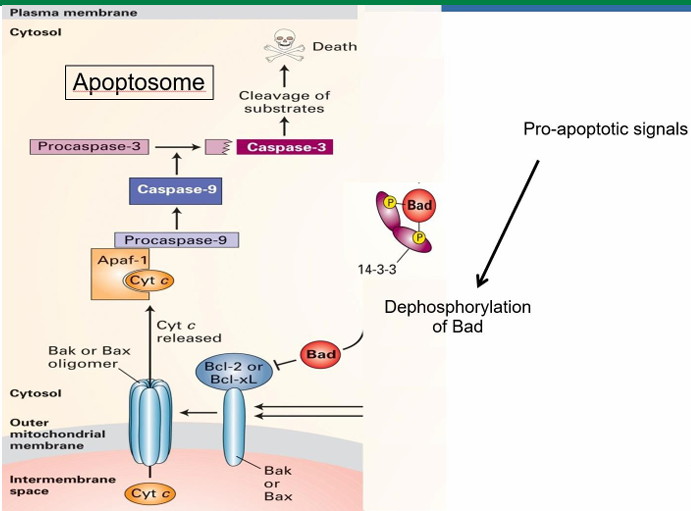
Trophic Factors in Apoptosis Prevention
Trophic factors prevent apoptosis to keep the cell alive
They initiate a kinase cascade leading to phosphorylation of the Bad protein
When trophic factors are removed, Bad can be dephosphorylated