TP53 and Apoptosis
1/25
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No study sessions yet.
26 Terms
What is the role of TP53 and which chromosome is it found on?
Chromosome 17
TF
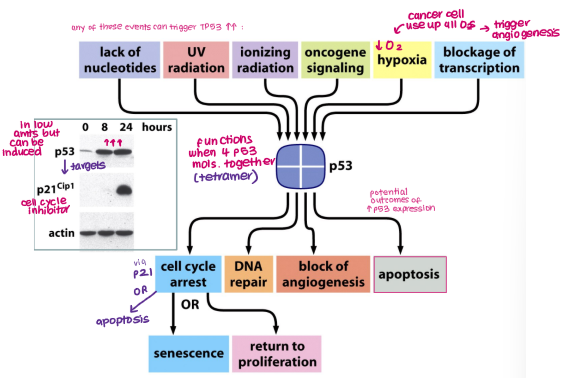
Guardian of the genome: responds to various events (e.g. hypoxia, DNA damage, oncogene signalling) and induces senescence, DNA repair, blocking angiogenesis and apoptosis
TP53 is triggered by certain events:
Hypoxia
DNA damage (UV radiation, Lack of nucleotides)
Oncogene signalling
Blockage of transcription
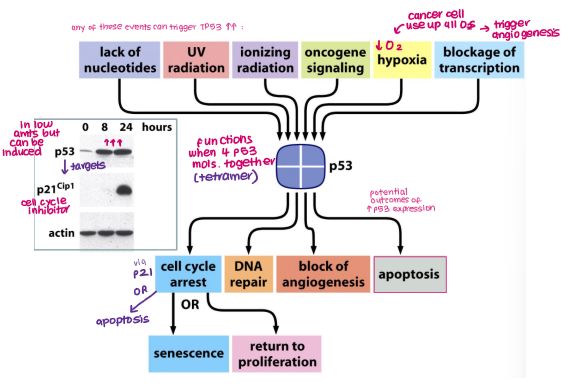
TP53 responds to events by inducing:
Senescence
DNA repair
Apoptosis
Blocking angiogenesis
Which two hallmarks of cancer is TP53 most implicated in?
Evading growth suppressors
Resisting apoptosis
TP53 mutations cause which syndrome? Which cancer is this implicated in?
Li-Fraumeni syndrome
Many cancers (e.g. ovarian, colon)
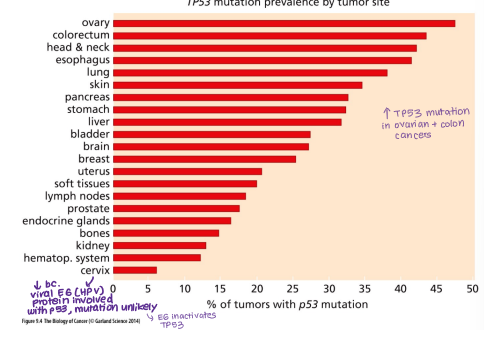
Why is TP53 mutation not commonly seen in cervical cancer?
HPV E6 protein already inactivates TP53
What is the structure of the TP53 gene like?
DNA binding domain (most mutations happen here)
NLS (TP53 is TF so stays in nucleus)
Mdm2 binding site (Mdm2 binds and degrades TP53)
Tetramerization site
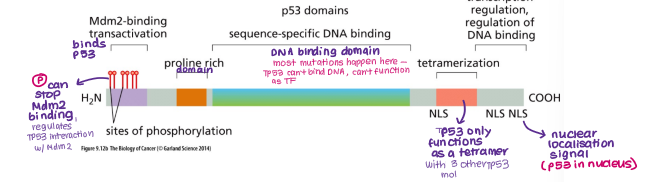
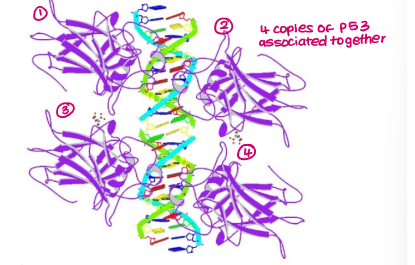
For TP53 to function, what needs to happen first?
forms tetramer with 3 other TP53 molecules
TP53 regulates expression of different genes. Give examples.
Mdm2
Growth arrest genes
DNA repair genes
Regulators of apoptosis (PTEN, BCl2, Bax)
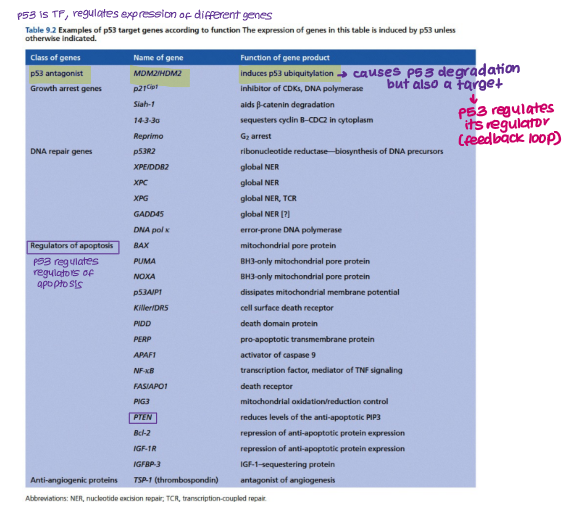
Which protein regulates TP53?
Mdm2 (negative regulator of TP53) BUT TP53 also regulates Mdm2 in a negative feedback loop
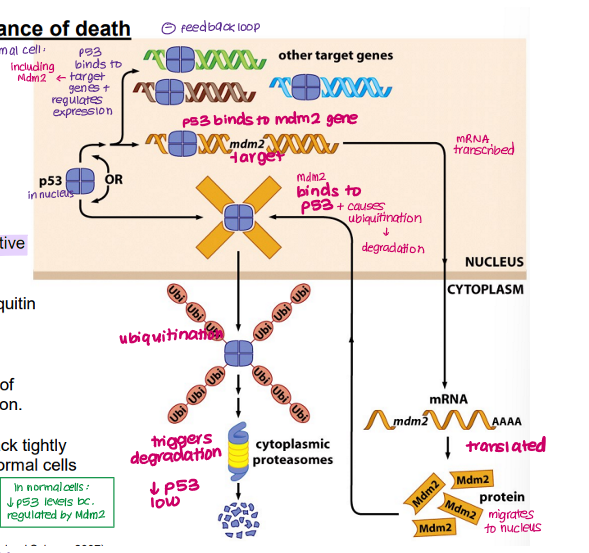
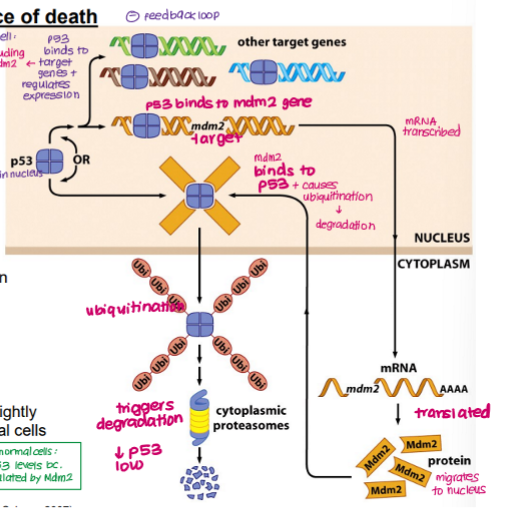
Explain how the balance between TP53 and Mdm2 is maintained in a normal cell.
TP53 binds and regulates expression of target genes (e.g. Bad, BCl2) via DNA binding domain
TP53 also binds Mdm2, increasing expression
MdM2 → transcribed and translated → Mdm2 protein
Mdm2 protein migrates to nucleus, binds TP53
TP53 ubiquitinated and degraded (prevents accumulation of TP53)
LOW TP53
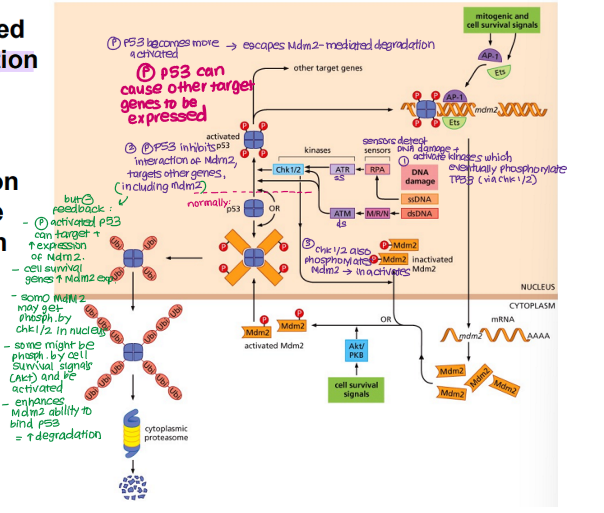
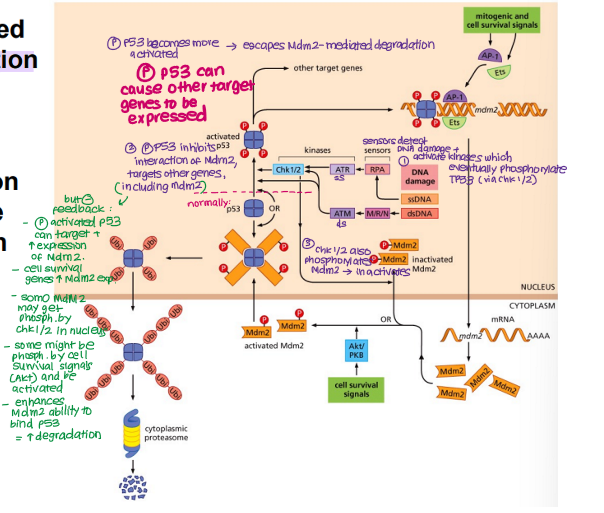
Explain how the balance between TP53 and Mdm2 is maintained in a normal cell when DNA damage occurs
TP53 phosphorylated in response to DNA damage
DNA damage/stress activates kinases which activate other kinases
Chk1/2 phosphorylates TP53 → ACTIVE
Phosphorylated/activated TP53 inhibits Mdm2 binding → no degradation
Chk1/2 phosphorylates Mdm2 → INACTIVE
TP53 accumulates → HIGH TP53
TP53 binds and induces expression of target genes (e.g. DNA repair genes, pro-apoptotic genes and Mdm2 itself)
Akt survival signals phosphorylate Mdm2 → ACTIVE
Active Mdm2 binds TP53 → ubiquitination → degradation → LOW TP53
Give two factors that affect Mdm2/TP53 interaction?
Phosphorylation
Cell cycle progression
Phosphorylation __________ TP53 activity and ___________ Mdm2 activity
Increases TP53 activity
Decreases Mdm2 activity (via Chl1/2) and increases activity via Akt surviv
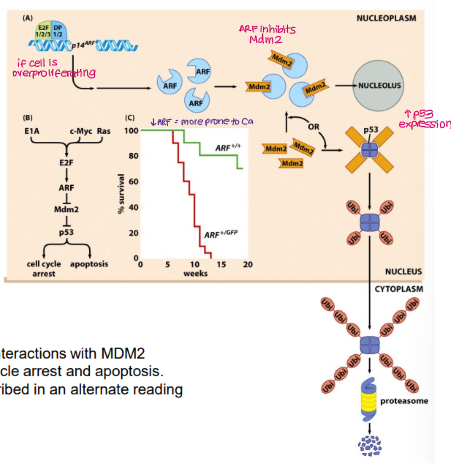
How does cell cycle progression affect MDM2 and TP53?
When cells proliferate → Rb inhibited → E2F activated
E2F drives expression of:
genes needed for G1→S phase progression
ARF tumour suppressor
ARF inhibits Mdm2 → can’t degrade TP53 → increases TP53
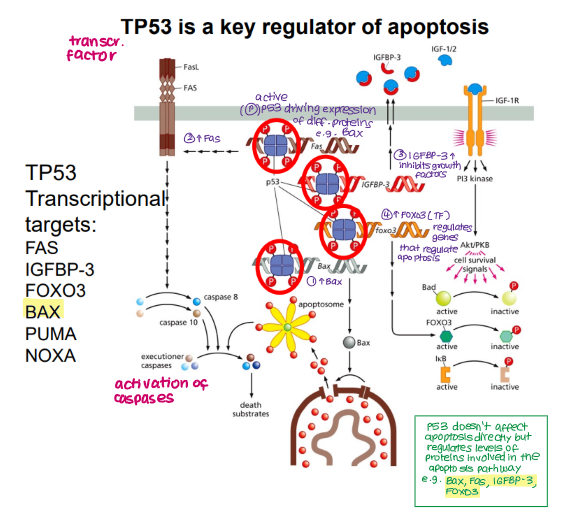
How does TP53 affect apoptosis?
Doesn’t affect apoptosis directly but regulates levels of proteins involved in the apoptotic pathway
Give examples of proteins involved in apoptosis regulated by TP53
Fas (extrinsic)
Bax BH123 protein (intrinsic)
IGFBP-3
FOXO3
PUMA
NOXA
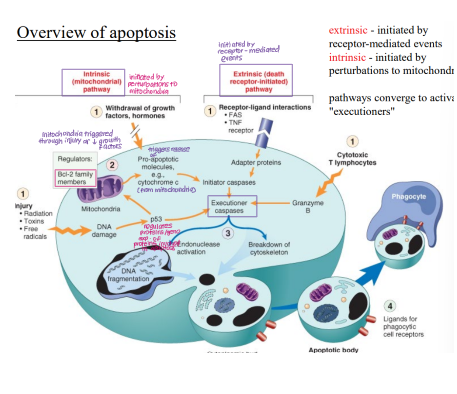
What are the two apoptotic pathways mediated by and where do they converge?
Extrinsic (receptor mediated, e.g. Fas death receptor, TGF receptor)
Intrinsic (mitochondria mediated, CD8+ T cells, cell injury, lack of growth signals)
Converge at proteolytic executioner caspases
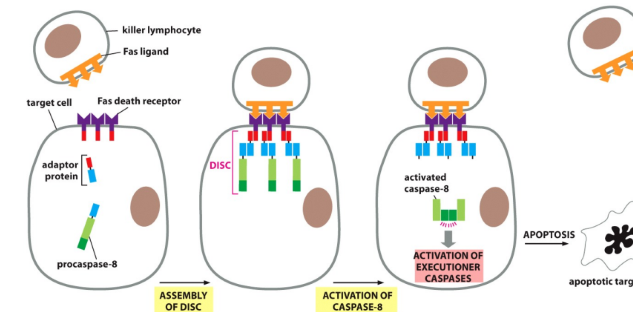
Describe the extrinsic apoptotic pathway and TP53 involvement
TP53 increases death receptor expression (Fas)
Fas ligand binds to Fas death receptor
DISC (death induced silencing complex) forms
Activates procaspases → caspases → executioner caspases
Describe the intrinsic apoptotic pathway and TP53 involvement
TP53 induces expression of pro-apoptotic genes (BH123 and BH3-only proteins) and inhibits expression of anti-apoptotic BCl2 genes
Mitochondria releases cytochrome C
Activates caspases → executioner caspases
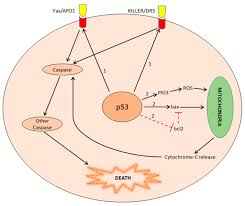
Intrinsic apoptotic pathway is regulated by
BCl2 family proteins
BCl2 proteins → ANTI-apoptotic
BH123 proteins → PRO-apoptotic
BH3-only proteins → PRO-apoptotic
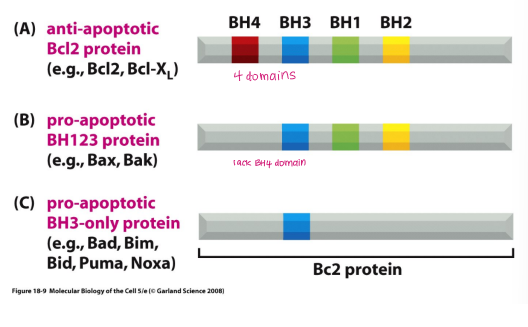
Give an example of BH123 proteins
Pro-apoptotic
Bax
Bak
Give an example of BH3-only proteins
Bad
Bid
Bim
Puma
Noxa
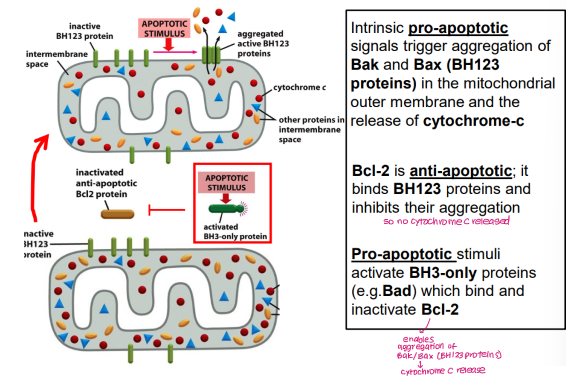
How do Bcl2 proteins regulate apoptosis?
Intrinsic pro-apoptotic signals trigger aggregation of BH123 proteins Bak and Bax in the mitochondrial outer membrane, causing release of cytochrome C
Anti-apoptotic Bcl2 binds BH123 proteins and inhibits aggregation
Bcl2 is inhibited by BH3-only proteins (e.g. Bad, Bid, Bim, Puma, Noxa)
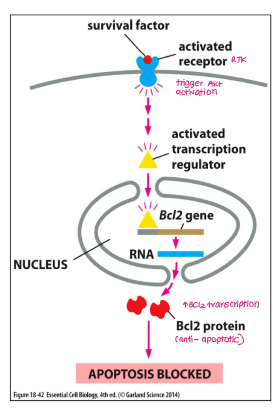
Bcl2 is regulated by
survival factors (repress apoptosis)
BH3-only proteins are regulated by
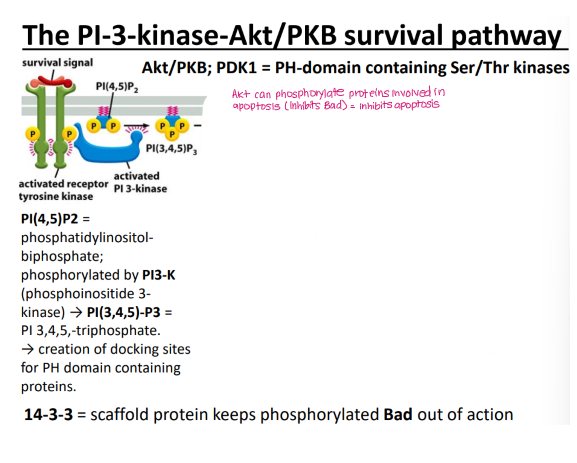
PI3K/Akt pathway
Akt inhibits Bad via 14-3-3 scaffold protein → inhibits apoptosis