
Looks like no one added any tags here yet for you.
Constitutively expressed genes
Always expresses
Regulated genes
Expressed only when needed, induced or repressed
Transcriptional regulation
Involves turning on/off genes at the level of RNA synthesis
Post-transcriptional regulation
Involves steps after RNA is made
Susceptibility of regulation through access to DNA by RNA polymerase
Not a large issue in bacteria, more significant in eukaryotes
Susceptibility of regulation through recognition of promoter
Mostly regulated by sigma factors in bacteria
Susceptibility of regulation through initiation
Even if sigma-RNA pol recognizes the promoter, sometimes other proteins are needed to enhance initiation. Other proteins may block transcription from ignition to elongation
Susceptibility of regulation through elongation
Some proteins slow down elongation, or cause premature termination
Susceptibility of regulation through termination
Sometimes termination can be overridden by anti-terminator protein
How might promoter sequence impact RNA polymerase?
Different promoters have different strengths
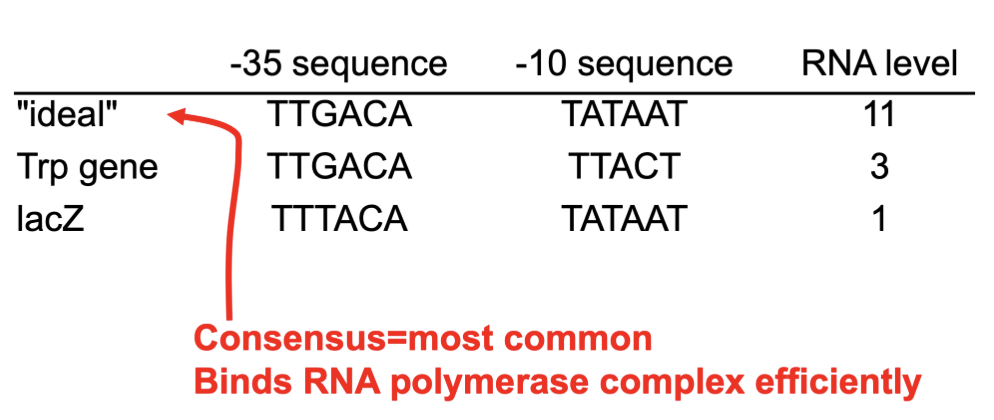
These sequences bind sigma factors
Overriding Attenuation of trp operon
trp operon: a cluster of genes in bacteria that encode enzymes involved in the biosynthesis of the amino acid tryptophan
attenuation occurs as long as tryptophan is plentiful
If tryptophan is low, ribosomes stall, preventing the formation of a hairpin, where there is typically attenuation
Cis-acting elements
DNA sequences located near a gene that directly regulate its expression
trans-acting elements
DNA sequences encoding upstream regulators (ie. trans-acting factors), which may modify or regulate the expression of distant genes. Trans-acting factors interact with cis-regulatory elements to regulate gene expression.
Lac operon
first operon discover
lacY: Galactoside permease (transports lactose into cells)
lacZ: B-galactosidase (cuts lactose into galactose and glucose)
lacA: Galactoside transacetylase (function unclear)
all 3 genes transcribed together producing 1 mRNA
has its own ribosome binding site
Can be translated by separate ribosomes that bind independently of each other
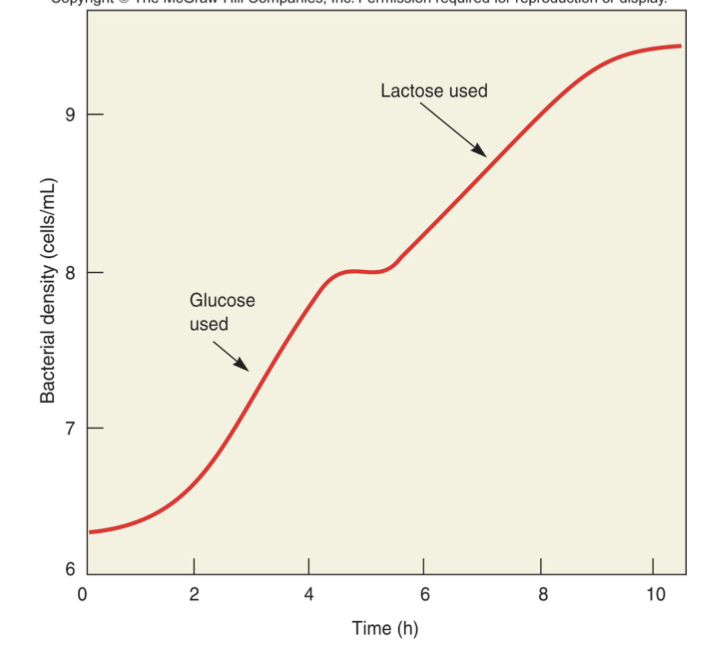
Diauxic growth of E. coli
two-phase growth pattern that occurs when microorganisms are given two sources to metabolize
cells are grown in both glucose and lactose
Lactose
Disaccharide, galactose and glucose joint by a b-galactosidic bond
needs to be imported into E. coli and digested into a monosaccharide
Negative Control of the lac operon
Lac repressor must be removed
where a repressor protein, encoded by the lacI gene, binds to the operator region on the DNA, preventing transcription of the lac operon genes unless lactose (the inducer) is present
Minor metabolic product of lactose, allolactose, is the inducer, removing tetramer and allowing transcription of b-galactosidase to break down lactose
IPTG (Isopropylthiogalactoside)
Analog for Allolactose (inducer), but cannot be broken down as an energy source, so it keeps lac operon “on”
colonies that are on are blue because it is producing b-galactosidase
Positive controller of lac operon
cAMP
A protein factor:
Catabolite activator protein (CAP)
Cyclic-AMP receptor protein (CRP)
gene encoding this protein is crp
Catabolite activator protein
cAMP added to E. coli can over come catabolite repression of the lac operon
addition of cAMP leads to activation of the lac gene even in the presence of glucose
CAP-cAMP complex
Positively controls activity of b-galactosidase
CAP binds to cAMP tightly
mutant CAP not bind cAMP tightly
Compare activity and production of B-galactosidase using both complex
low activity with mutant CAP-cAMP
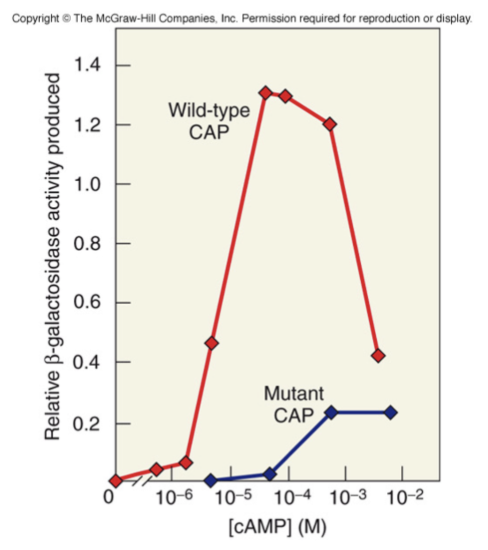
How does CAP-cAMP complex positively regulate lac operon
CAP-cAMP complex allows the formation of the open promoter complex for the RNA polymerase
proposed:
CAP-cAMP dimer binds to its target site on the DNA
alpha CTD of polymerase interacts with the specific site on CAP
Binding is strengthened between promoter and polymerase
ara operon use
Coded for enzymes required to metabolize the sugar arabinose
catabolite-repressible operon
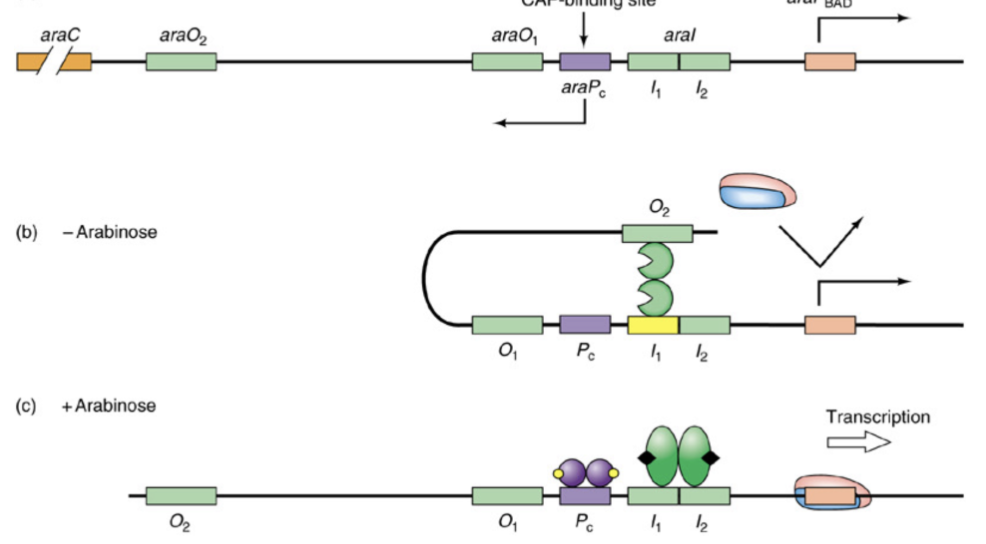
Structure of ara operon
3 genes (araB, A, and D) encode for arabinose metabolizing enzymes
transcribed rightward from promoter araPbad
Other gene, araC
encodes the control protein AraC
Transcribed left ward from the araPc promoter
Control of ara Operon
When lacking arabinose, a loop is formed that blocks binding site
RNA polymerase just bounces off
When there is arabinose, binding to araPc is able to occur and transcription of araBAD can occur
trp operon
Contains the gene for the anabolic enzymes (build up a substance) the bacterium needs to make the amino acid tryptophan
turned off by a high level of the substance provided
operon is subject to negative control by a repressor when tryptophan levels are elevated
negative control of trp operon
5 genes code for polypeptides in the enzymes of tryptophan synthesis
high tryptophan concentration signals to turn off the operon
the presence of tryptophan helps the trp repressor bind to its operator
No tryptophan: No trp repressor, just inactive protein aporepressor
If aporepressor binds tryptophan, changes conformation with high affinity for trp operator → forms trp repressor→ tryptophan is a corepressor
Riboswitches
Small molecules can act directly on the 5’ UTRs of mRNAs to control their expression