DNA Damage & Repair
1/37
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
38 Terms
Endogenous Damage
from internal environment
DNA Damage
chemical changes to the N-rich bases
can lead to changes in base pairing potential → mutation
disrupts flow of info from that site in the genome
can lead to double strand breaks → genome instability
can be repaired or cells undergo programmed cell death
Spontaneous Deamination
non-catalyzed (spontaneous) deamination of cytosine converts this base into uracil
uracil chemical is very similar to thymidine → base pairs with adenine
if left un-repaired, leads to A-T mutation
non limited to cytosine
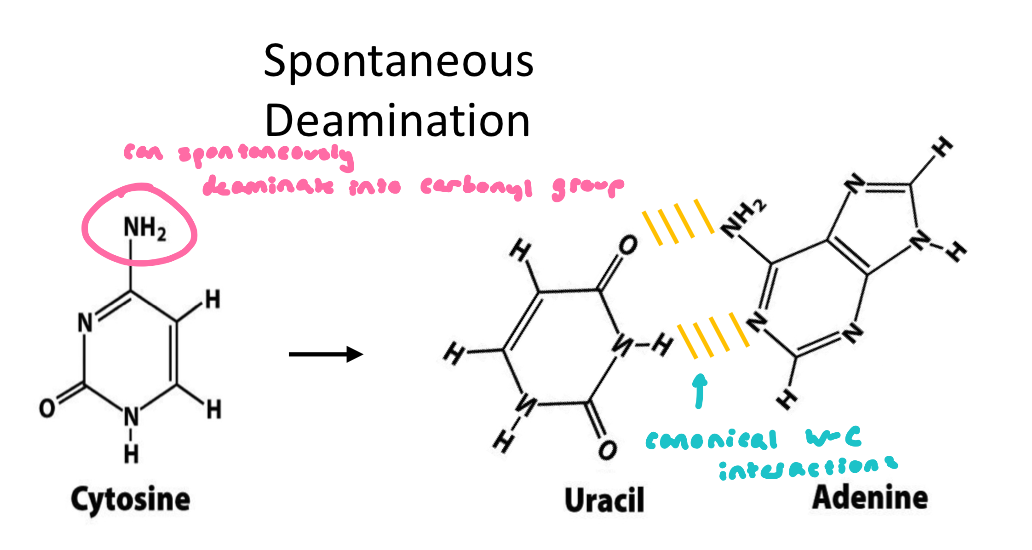
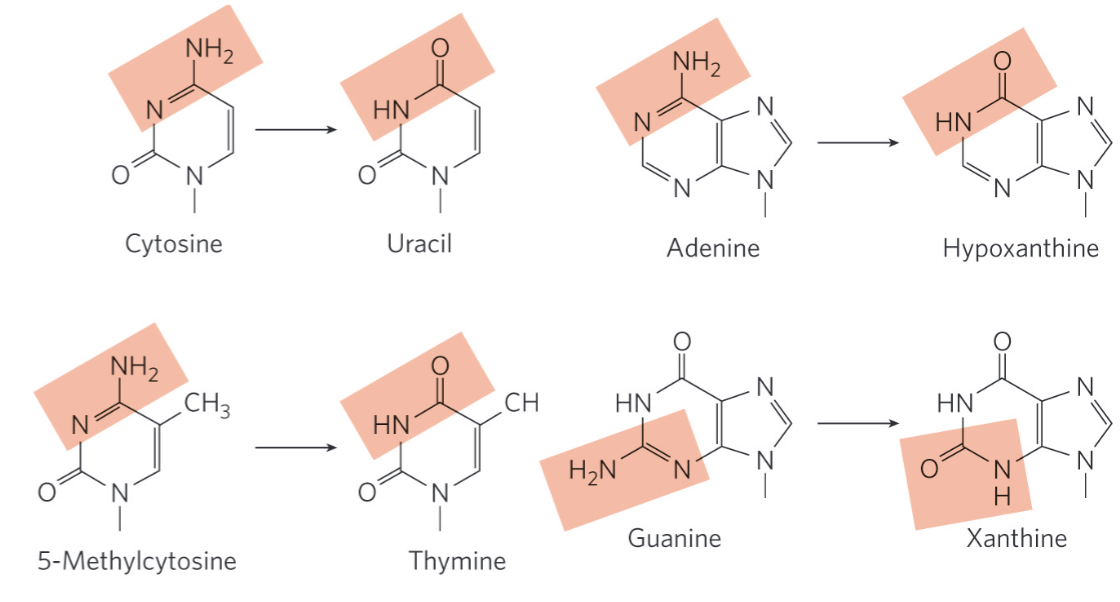
Deamination Reactions
deamination = spontaneous loss of exocyclic amino groups
deamination of cytosine to uracil = ~100 events/day
recognized as foreign (damage) in DNA and removed → why we don’t see uracil in the genome, and why DNA only contains thymine
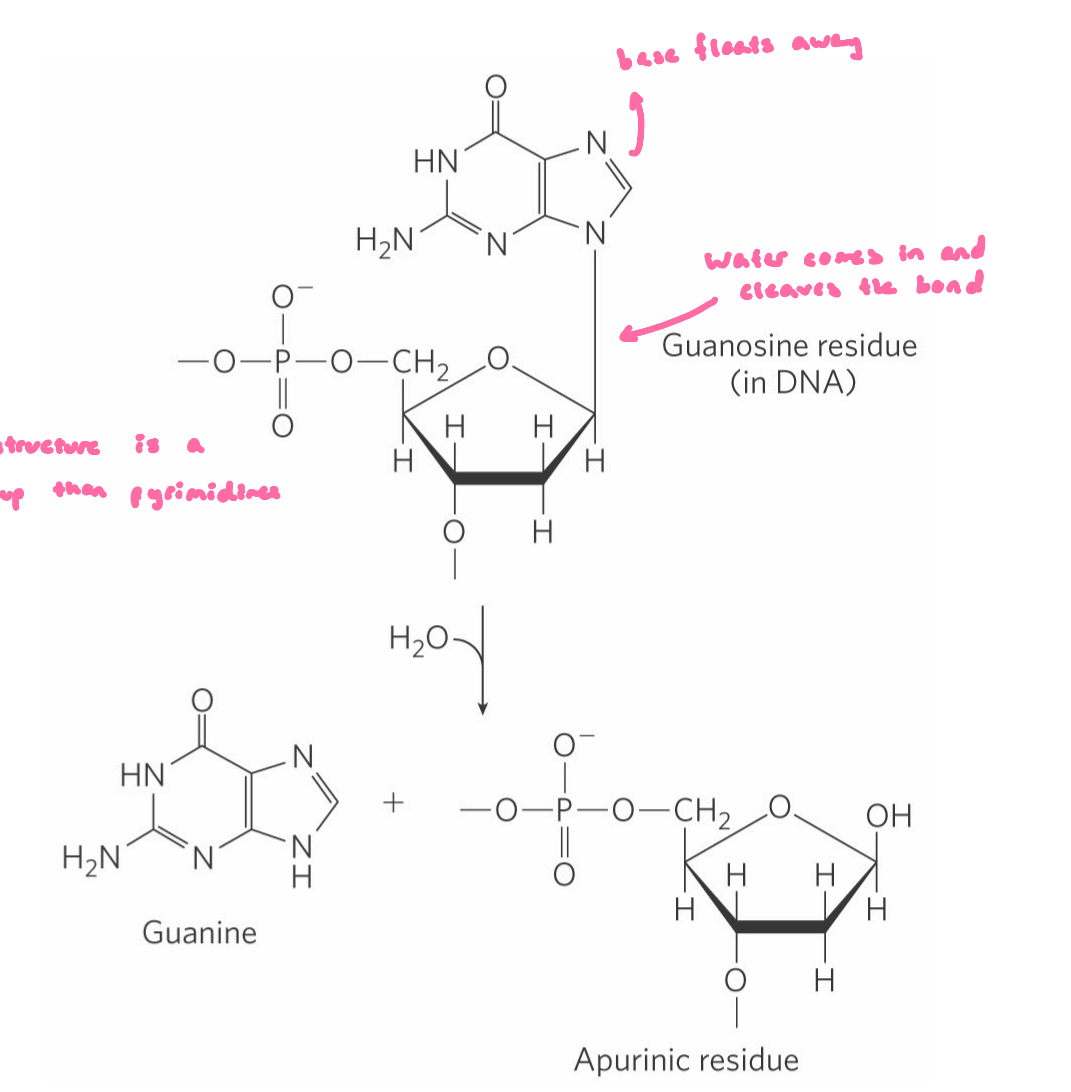
Depurination Reactions
depurination = hydrolysis of the N-β-glycosyl bond b/w the base and the pentose
creates an AP (apurinic, apyrimidinic) site or abasic site; a site with no information
more common with purine: double ring structure is a better leaving group than pyrimidines
polymerase doesn’t know what H-bonds will be made
leads to mutations, replication stalling, double-stranded breaks
DNA Damage by Oxidative Damage
reactive oxygen species (hydrogen peroxide, hydroxyl radicals, superoxide radical) damage DNA
radicals really want to bind to something → if they bind to an N-rich base and add an oxygen, it changes how that base can interact w/ other nucleotides
hydroxyl radicals are responsible for most oxidative DNA damage
cells have an elaborate defense system to destroy reactive oxygen species
as we get older, cells have more chances to accumulate reactive oxygen species
mitochondria leak more + release damaging oxygen molecules
DNA Damage by Oxidative Damage Figure
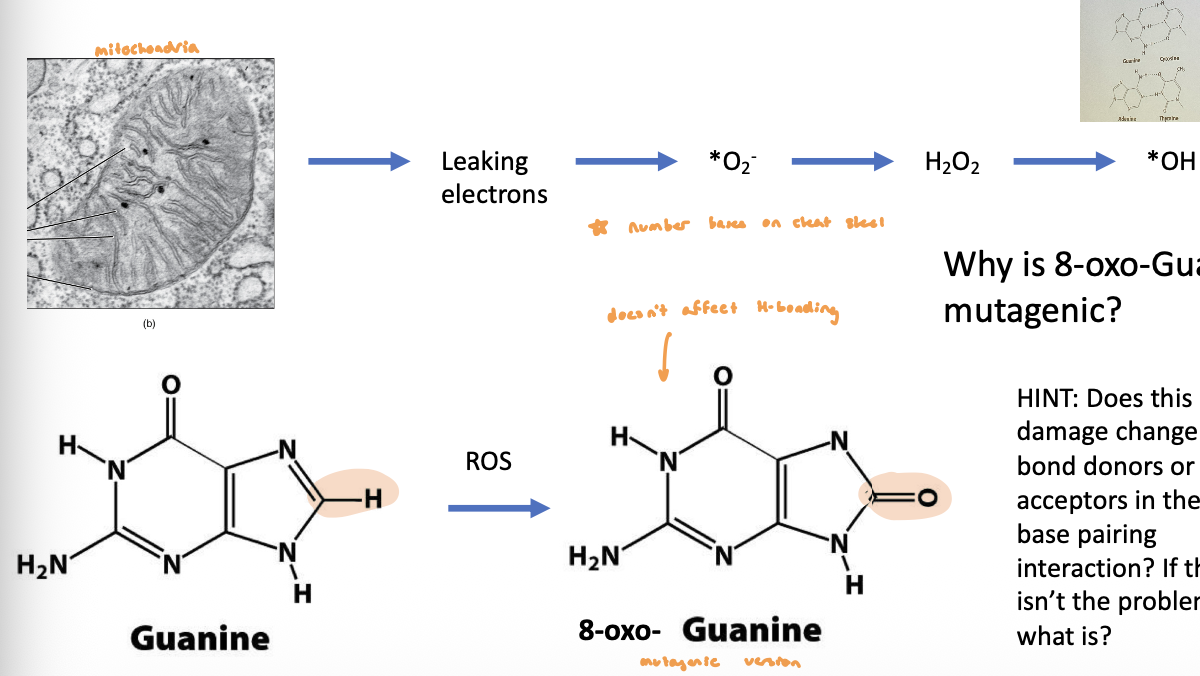
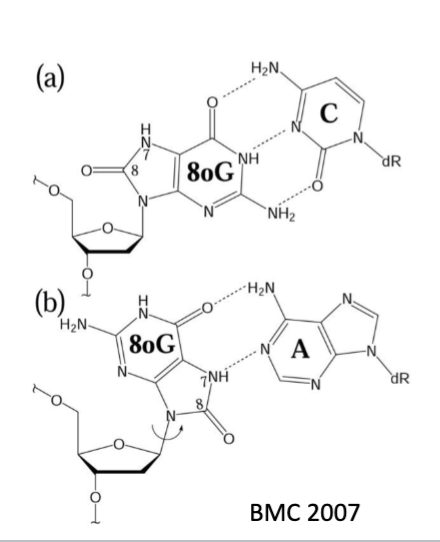
8-oxoG pairing
2 conformations of base and ribose (syn and anti)
anti conformation is usually favored, but new steric clash b/w carbonyl and ribose oxygen now increases Hoogsteen base pairin b/w G and A during replication
after a 2nd round of replication, this will generate an AT pair
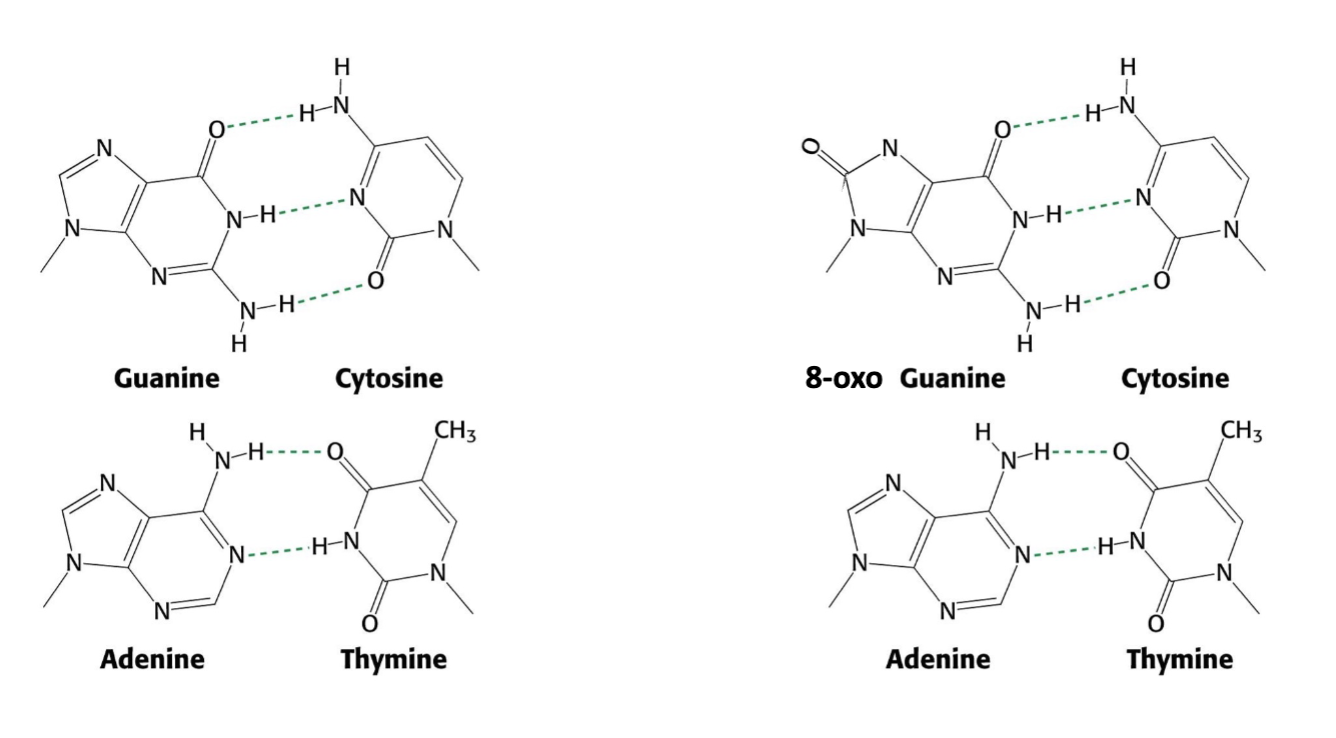
8-oxoG and A base pairing
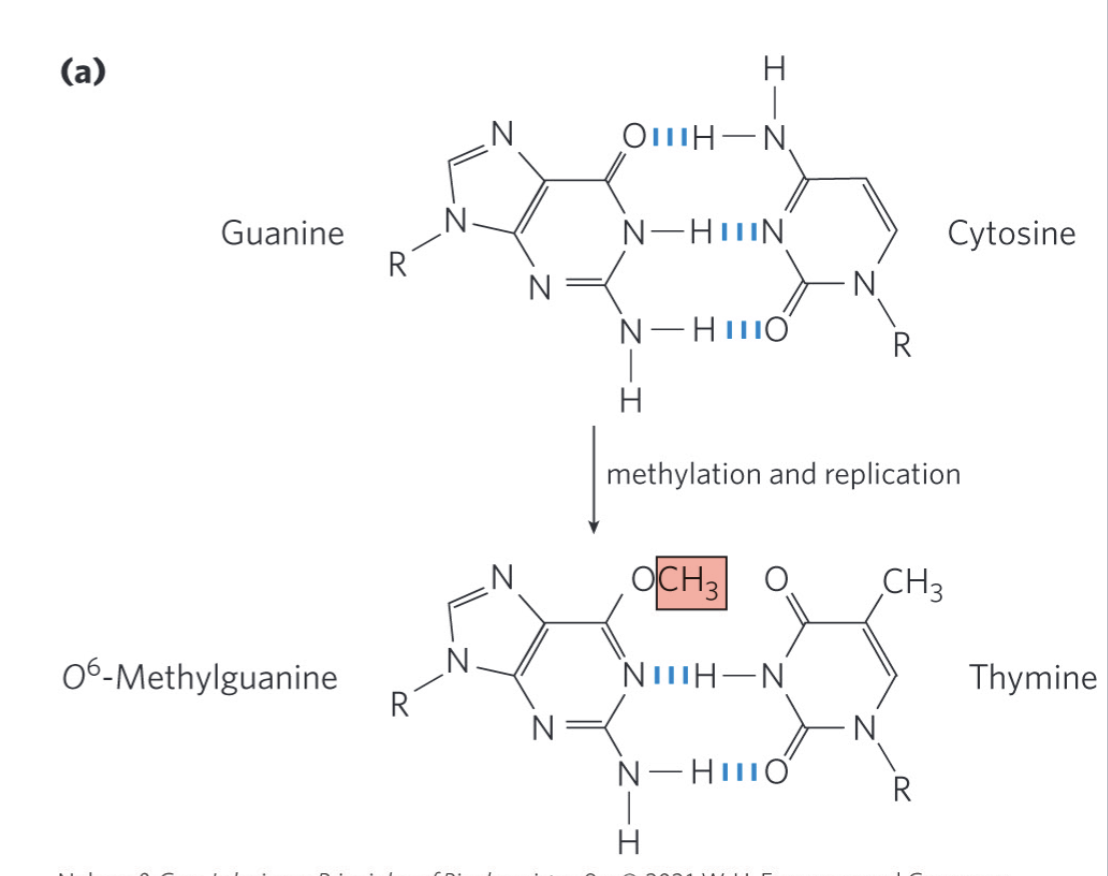
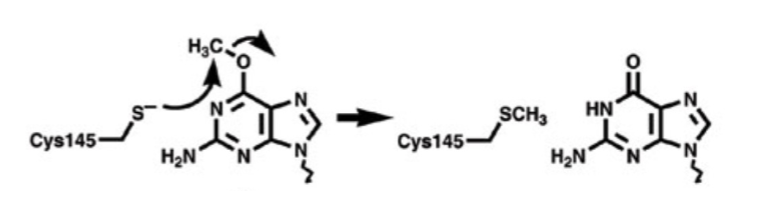
O6 Methylguanine DNA Methyltransferase
catalyzes transfer of the methyl group of O6-methylguanine to one of its own Cys residues
a single methyl transfer event permanently methylates the protein, inactivating
damage does not lead to mutations until after replication
Exogenous Damage
occurs from external environmental factors
changes ability of N-rich bases to make canonical W-C interactions
can lead to mutation after DNA replication
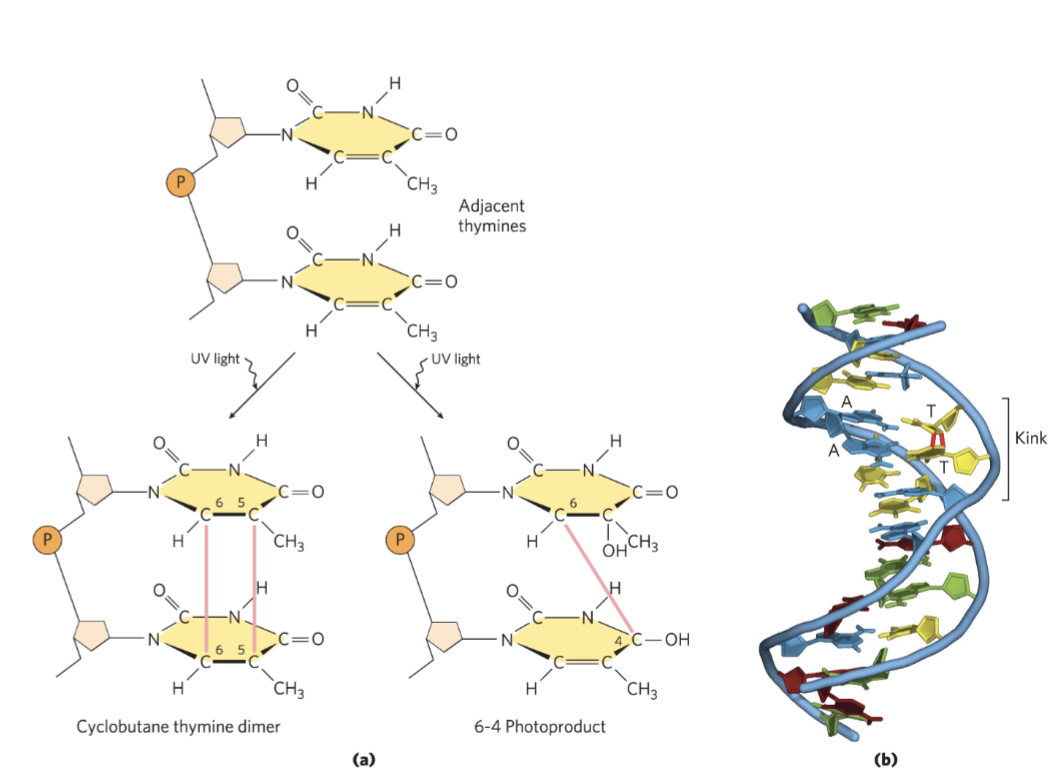
Pyrimidine Dimers promoted by UV Radiation
UV light causes pyrimidine dimers (2 thymines 2 cytosines in one row on one strand)
cyclobutane pyrimidine dimers (2 covalent bonds) or 6-4 photoproduct (1 covalent bond)
have the right geometry for covalent bonds b/w dimers when high energy radiation occurs in cell
UV damage can cause photo-crosslinking (occurs b/w pyrimidine dimers)
ionizing radiation (x-rays and gamma rays) causes: ring opening, base fragmentation, breaks in the covalent backbone of nucleic
Translesion Polymerase η Eta
if the covalent bonds have formed, H-bonds b/w bases can’t form
Eta is a translation polymerase that doesn’t repair UV-photo-cross linked DNA, it bypasses the lesion during replication by inserting the correct bases
larger active site fits the distorted cross linked T’s
no editing site
is not reading the template (it’s not readbale)
usually adds two dAs to UV damage, and usually adds a dC if it encounters 8-oxoG
then falls off → replication polymerase synthesizes the rest of DNA → prevents stalling
Foromation of Nucleotides w/ Alkylation Damage
O6-methylguanine = a modified nucleotide that forms in the presence of alkylating agents
common and highly mutagenic lesion
tends to pair w/ thymine rather than cytosine
if left unrepaired, T will base pair w/ A in the next cycle (AT mutation)
eg. cigarette smoke, moldy peanuts, burnt meat
Endogenous vs Exogenous Damage
Endogenous: spontaneous deamination, depurination, oxidative damage from respiration, DNA replication (polymerase errors)
Exogenous: photo-cross linking from UV (most common), chemical exposure (alkylation)
both types of DNA damage leads to heritable mutations after replication
Mismatch Repair
DNA Polymerase Errors
DNA polymerase has high fidelity: exonuclease site, shape discrimination, mismatch repair pathways
some mutations sneak by from time to time, which lead to an error rate ~10-9
mistakes can be catastrophic if it leads to certain mutations
3 billion base pairs: ~60 mistakes/cell division → mutation
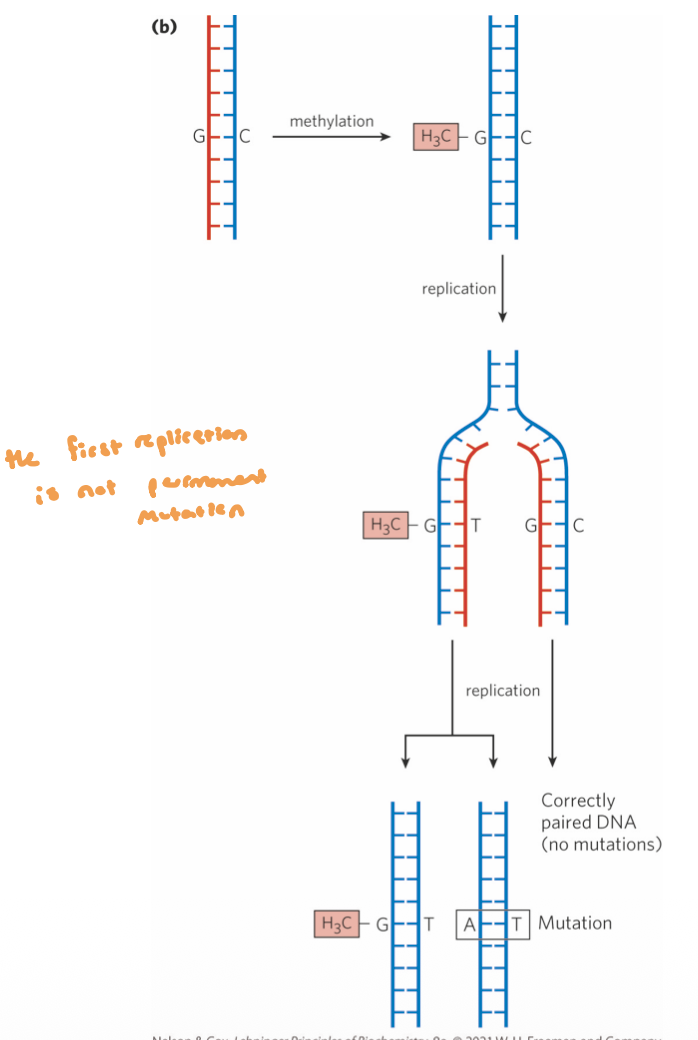
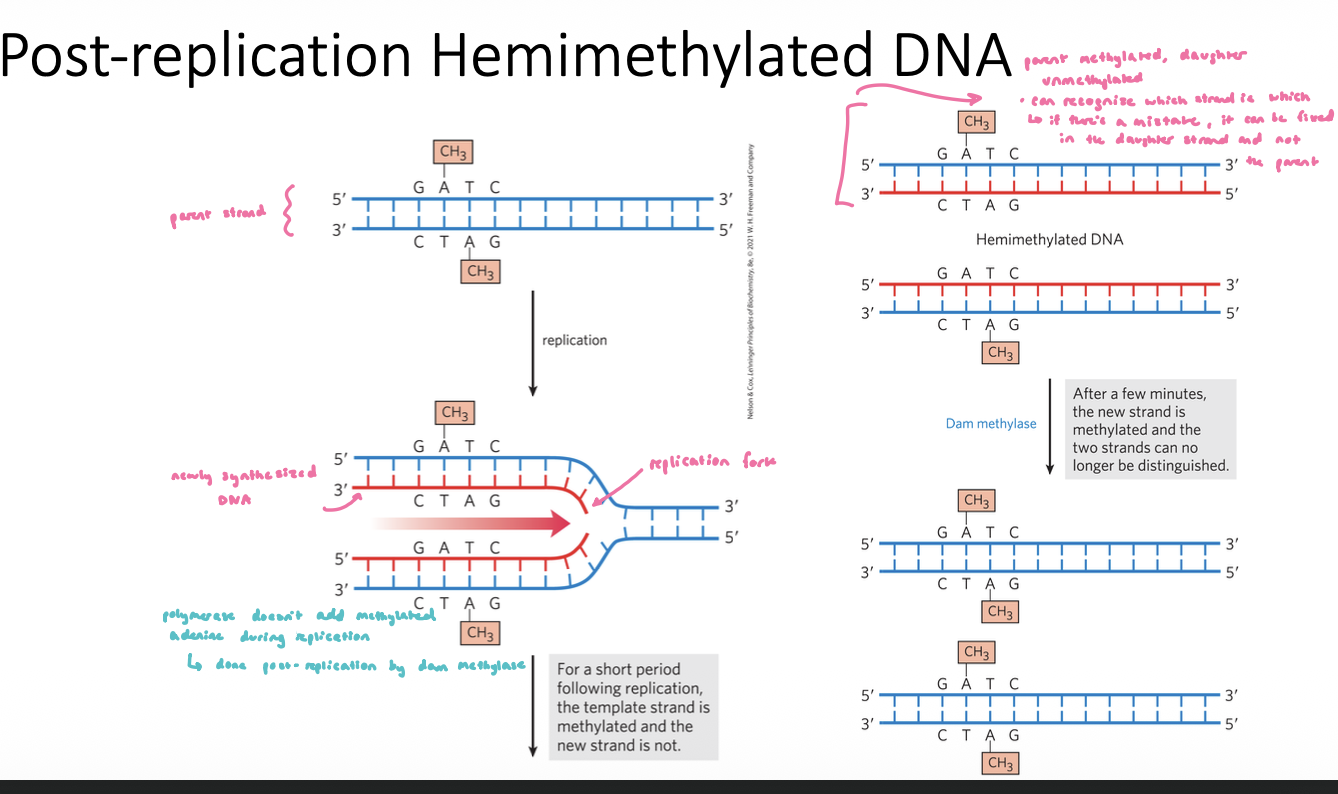
DNA Methylation in E.coli
cell can discern which is the parent strand and which is the daughter strand thru variations in DNA methylation
Dam Methylase (flagging system):
methylates GATC sites on the adenine residues
use the single-carbon transfer co-factor SAM
~1 min after replcation, this enzyme will methylate these sites in the newly synthesized daughter strand
follows replication closely, parent strand is methylated, daughter strand is unmethylated
this complex comes up and methylates the new daughter strand after replication
Post-Replication Hemimethylated DNA
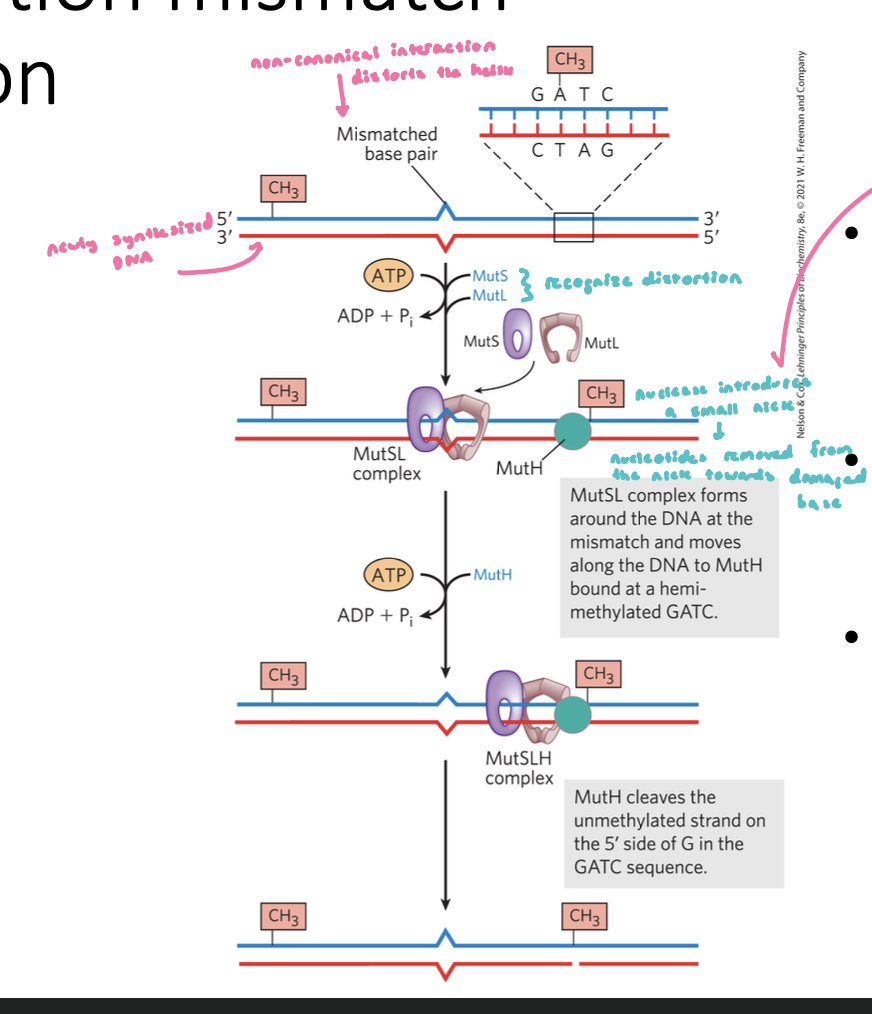
Post-Replication Mismatch Identification
non-canonical interactions distort the helix
MutS and MutL are loaded on to the dsDNA at the lesion (mismatch) in an ATP dependent manner
complex slides along the DNA until it encounters a GATC methylation site (GATC is used as a marker)
non methylated strand is cleaved generating a single-strand nick (nuclease)
nucleotides are removed from the nick towards the damaged base
nuclease also requires a helicase to unwind the DNA
Post-Replication Mismatch Identification FIGURE
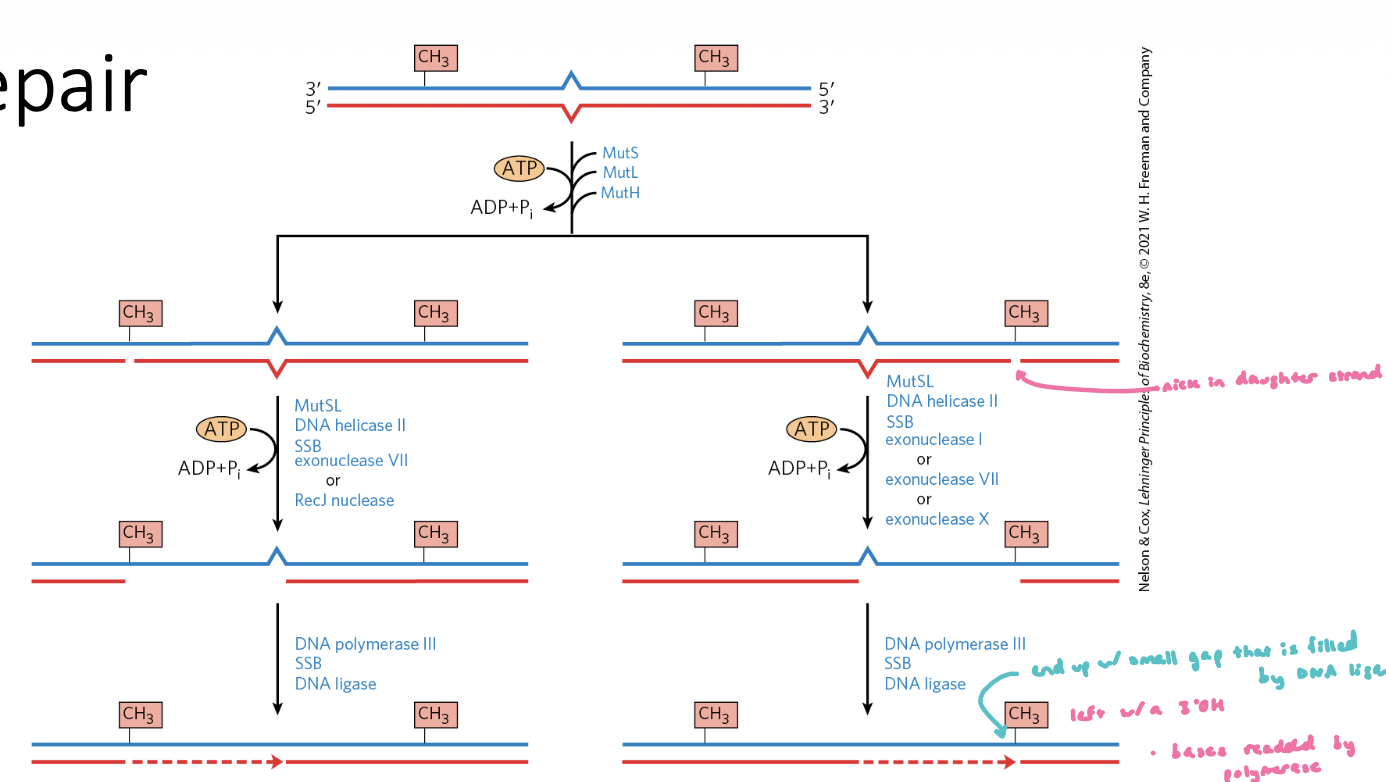
Mismatch Repair Steps
helicase unwinds the DNA at the nick site and moves towards the lesion
nucleotides b/w the nick and the lesion are removed by an exonuclease
nick or exonuclease activity generates a free 3’OH
non-damaged strand is used as a repair template for DNA polymerase
gap at the end of the repair is sealed w/ a ligase
dam methylase can methylate the GATC site
Mismatch Repair Steps FIGURE
Single-Strand Break Repair
damaged DNA can be repaired in different ways, depending on the type of damage (binding site for repair proteins to recognize)
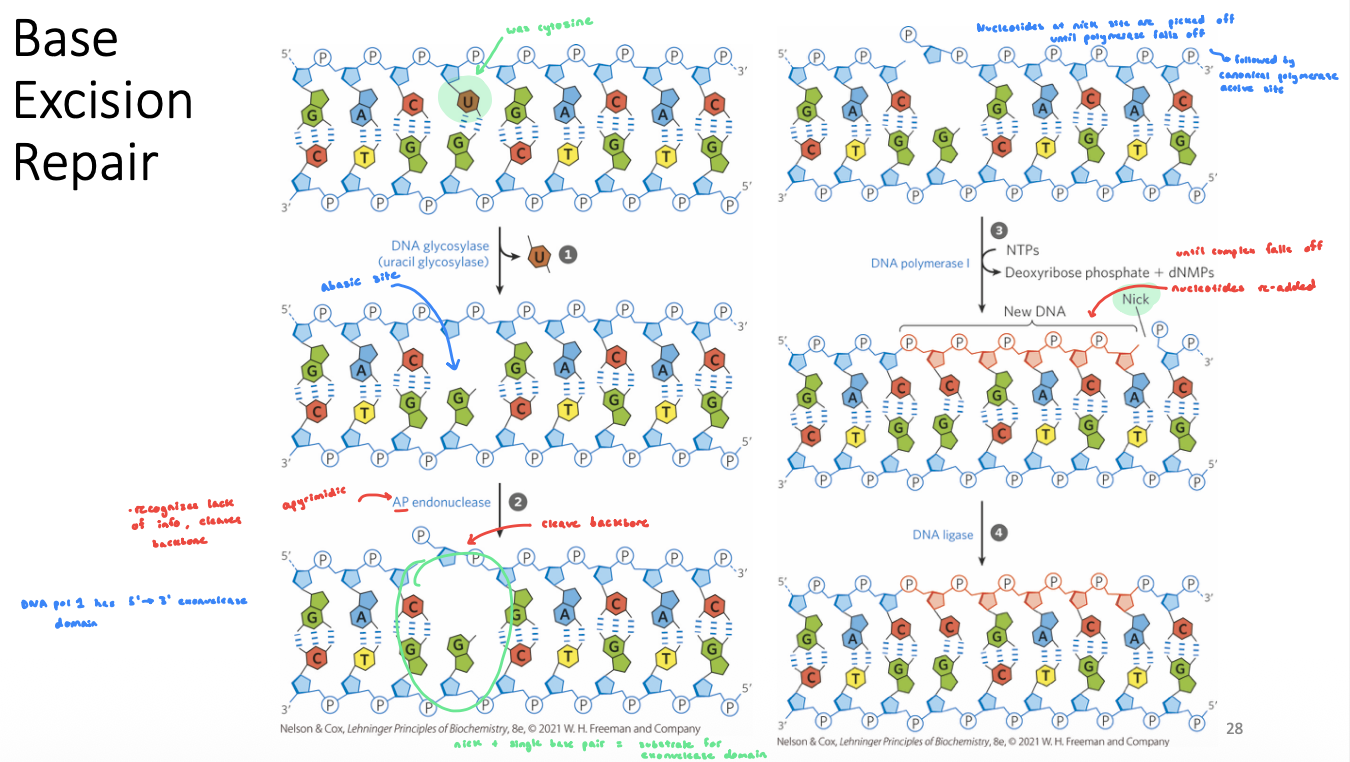
Base-Excision Repair (BER): Glycosylases
DNA glycosylases recognize common DNA lesions and remove the affected base by cleaving the N-glycosyl bond in BER → generates an abasic site
generally specific for one lesion type
uracil DNA glycosylases recognize uracil in DNA and remove them
BER Steps
damaged base is recognized by a specific glycosylase the removes the base (generates an abasic site which is a substrate for AP endonuclease)
an endonuclease cleaves the phosphodiester backbone at the abasic site
DNA Pol I replaces missing base (and short extension)
Gap generated by the polymerase extension is filled with DNA ligase
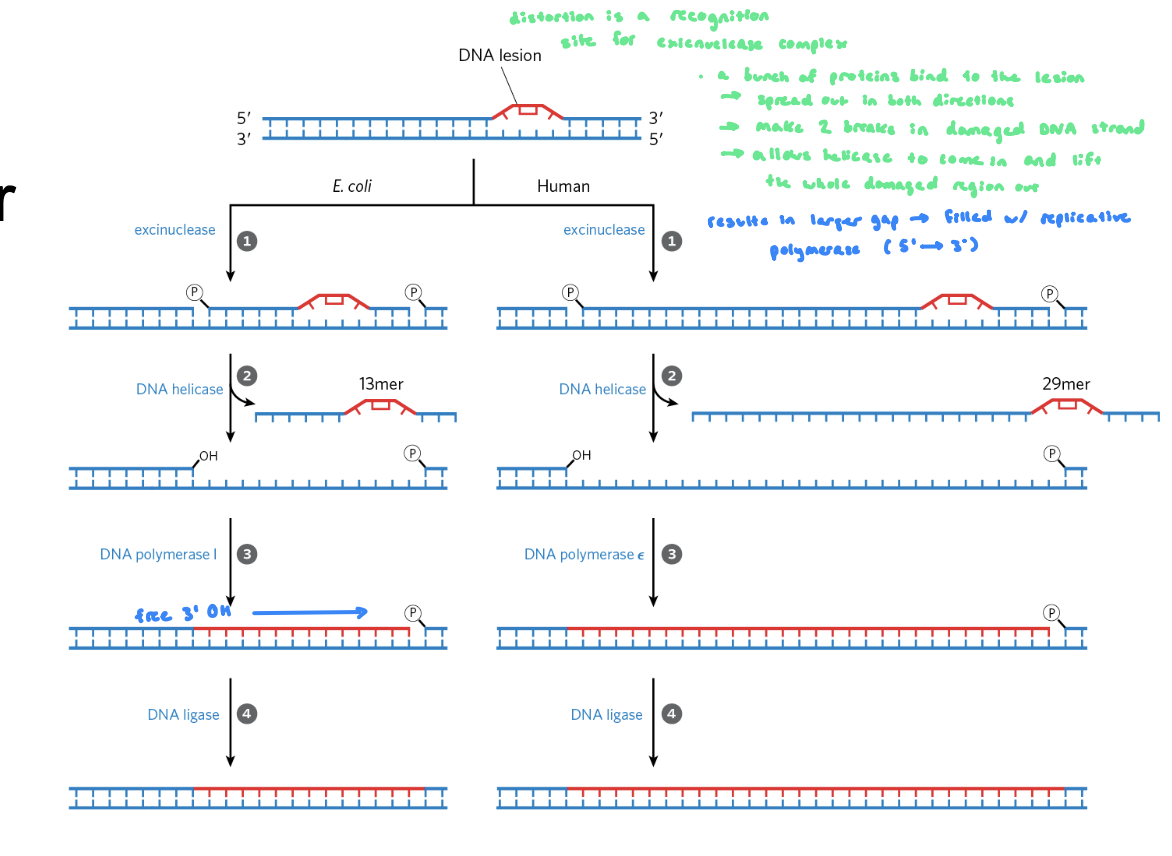
Nucleotide Excision Repair in E.coli and humans
occurs w/ larger distortions of DNA (e.g. photocrosslinking)
exicnuclease: a multisubunit enzyme that hydrolyzes two phosphodiester bonds, one on either side of the distortions
distortion is a recognition site for exicnuclease complex
a bunch of proteins bind to the lesion → spreads out in both directions → make 2 breaks in damaged DNA strand → allows helicase to come in and lift the whole damaged region out (flanking the damage site)
results in larger gap, filled w/ replicative polymerase (5’→3’)
DNA pol I (E.coli) or DNA pol ε (humans)
DNA ligase seals the nick
NER Figure
Direct Repair: MGMT
requires even more specificity
MGMT directly removes the damage (methyl-group) and the chemistry (binding potential) of the guanine residue is restored → restores to a canonical nucleotide
MGMT is unable to release methyl-group after binding and degraded after a single reaction
single turnover protein (not an enzyme)
if we want to do the rxn again, more protein has to be synthesized
costs the most ATP
MGMT
binds to alklyated guanine residues – flips the base out and removes the methyl group – restoring the guanine to canonical W-C binding potential and mitigating mutation risks
single turnover protein → costs a lot to make a protein
Direct Repair: MGMT FIGURE
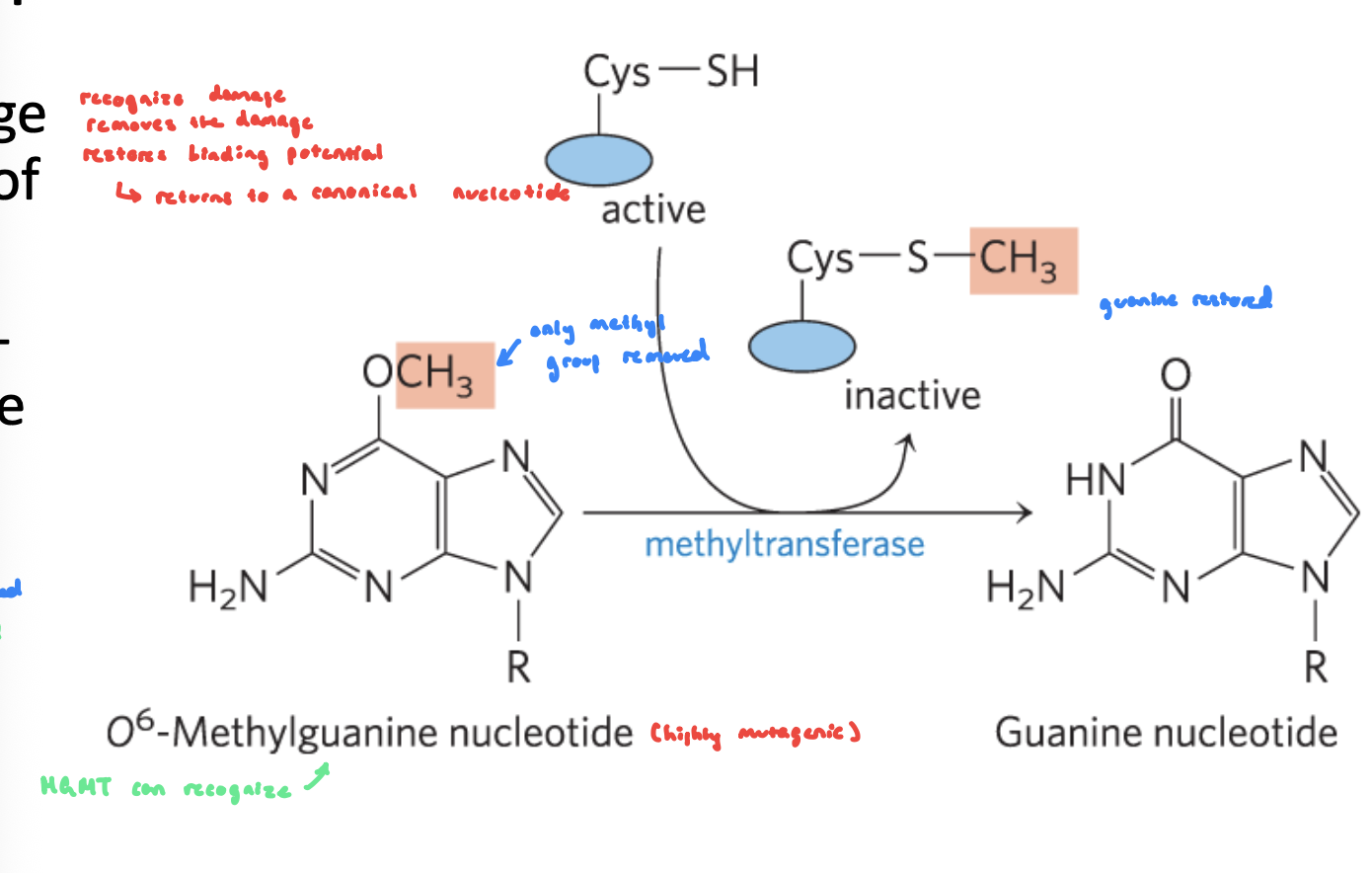
Role of Cystein in MGMT
Cys thiol group performs a nucleophilic attack on the alkyl group attached to the O⁶ position of guanine
The alkyl group is transferred from guanine to cysteine
irreversible reaction
Role of Arginine in MGMT
helps with DNA binding and lesion stabilization
Arginine is positively charged and DNA backbone is negatively charged
Helps position the damaged guanine correctly in the active site
May help flip the damaged base out of the helix (base flipping)
Cancer
damage by exogenous or endogenous sources cause lesions in DNA, if unrepaired these can lead to mutations after DNA replication
non-functional repair pathways can accelerate mutation, if these mutations disrupt the relationship b/w cell growth and division, this can lead to cancer
carcinogens have high capacity to damage DNA and drive mtations and can be detected using Ames test
Mutations are Linked to Cancer
mutation: a permanent change in the nucleotide sequence (in coding or non-coding regions)
substitution mutation: replacement of one base pair w/ another (missense mutation); changes a.a in protein
insertion mutation: the addition of 1+ base pairs; deletion mutation: the deletion of 1+ base pairs
caused by dsDNA repair pathways
silent mutation: a mutation that affects nonessential DNA or has negligible effect on gene function
in coding region, doesn’t change a.a. in protein, but can change its ability to bind to tRNA
nonsense mutation: a mutation tha causes a pre-mature stop codon (results in truncated protein)
it doesn’t require many mutation in a cell in order to drive cell to transformation (uncontrollable growth, lacks cues for growth, cell division/cycle)
takes very specific mutation
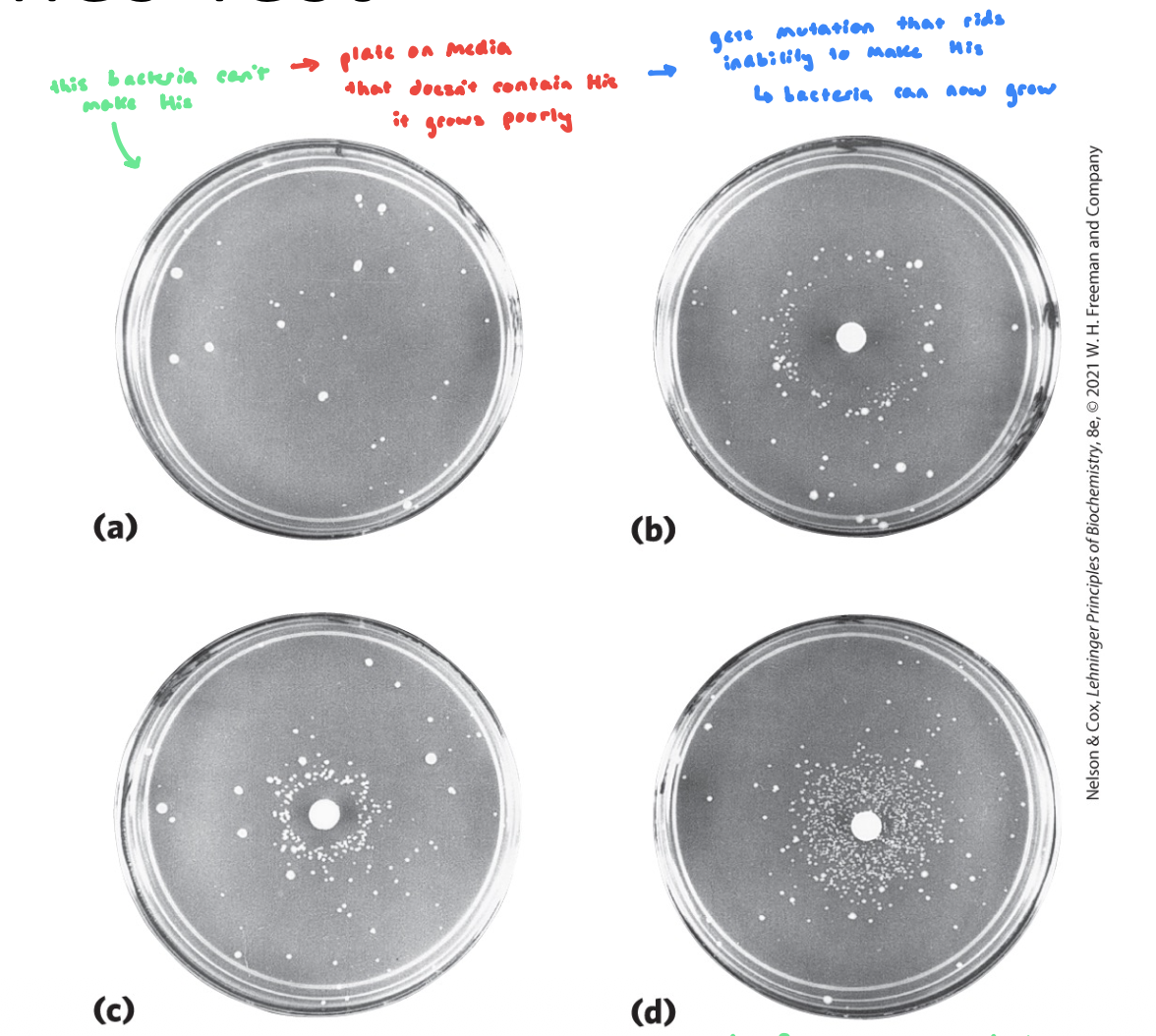
Ames Test
detects mutagenic compounds
Salmonella thpimurium w/ a mutation in the His synthesis pathway is grown on His-free plates (can’t make His)
a small circle of Whatman filter paper is soaked in a potential mutagen (ask if it drives mutations)
colonies will arise in mutagenic conditions, after the mutation relieves the histidine synthesis blockage
the further we get colonies, the more mutagenic (even at dilute conc’s we’re getting mutations)
Ames Test FIGURE
DNA Repair is Tumor Protective
cancer is driven by changes in genome of a cell that allows it to be oncogenic (often involves mutations in DNA repair genes)
DNA damage can be used as a cancer treatment: damages DNA at tumor site, if you get enough damage, it can drive cell to apoptosis