Dunn M&C - Prokaryotic Transcriptional Regulation
1/34
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
35 Terms
promoter
initiation of transcription; directs the transcription machinery to the 5’ end of a gene
terminator
termination of transcription; causes RNA polymerase to dissociate from the template DNA, thereby ending transcription
transcription factor
sigma factor — e.coli that is required for transcription
transcription three stages
initiation
elongation
termination
applies to both prokaryotes and eukaryotes
regulatory transcription factors
determine how efficiently (and therefore frequently) a transcriptional initiation complex forms and successfully escapes the promoter
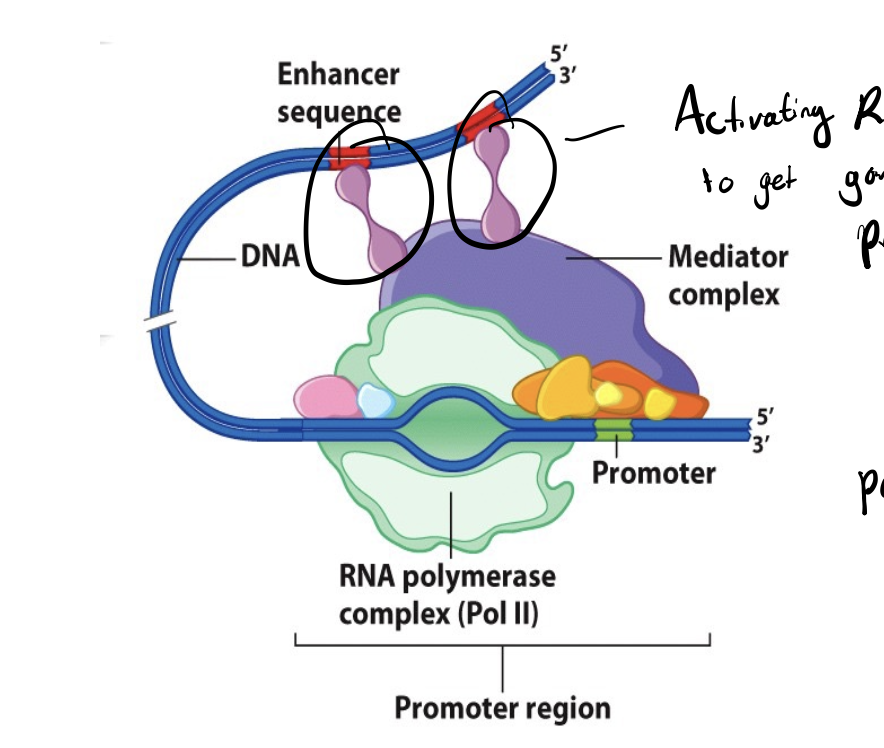
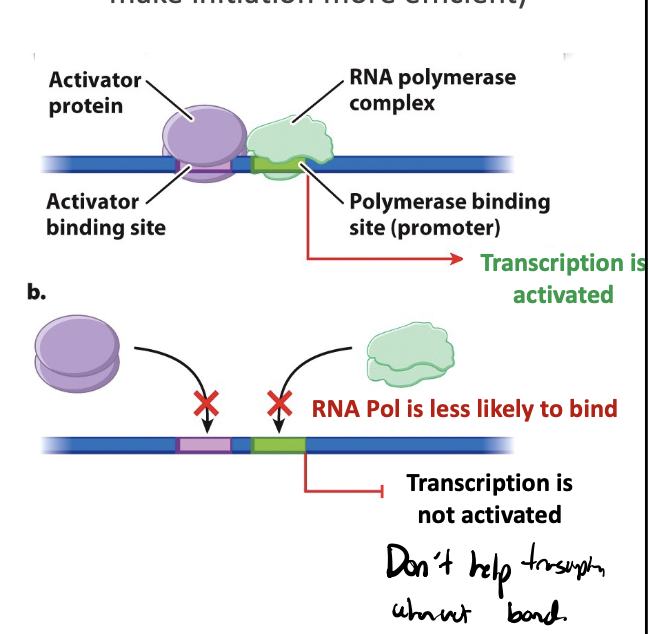
positive regulation
leads to transcriptional activators
transcriptional activators
protein that promotes transcription; helps recruit RNA polymerase or make initiation more efficient
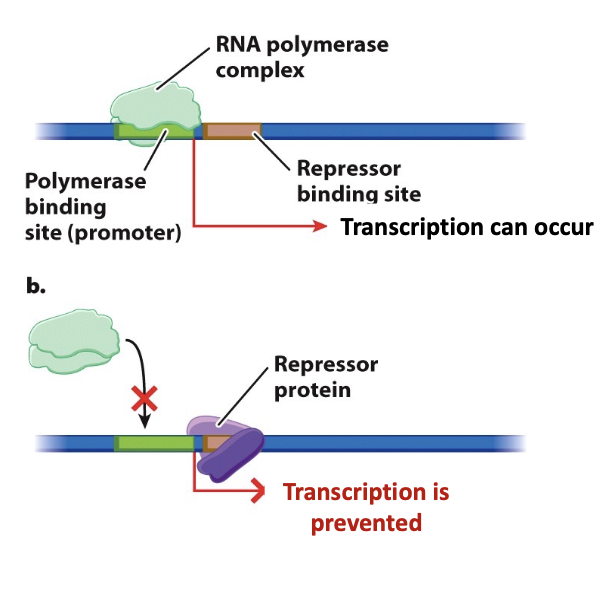
negative regulation
leads to transcriptional repressors
transcriptional repressor
protein that inhibits transcription; prevents recruitment of RNA polymerase
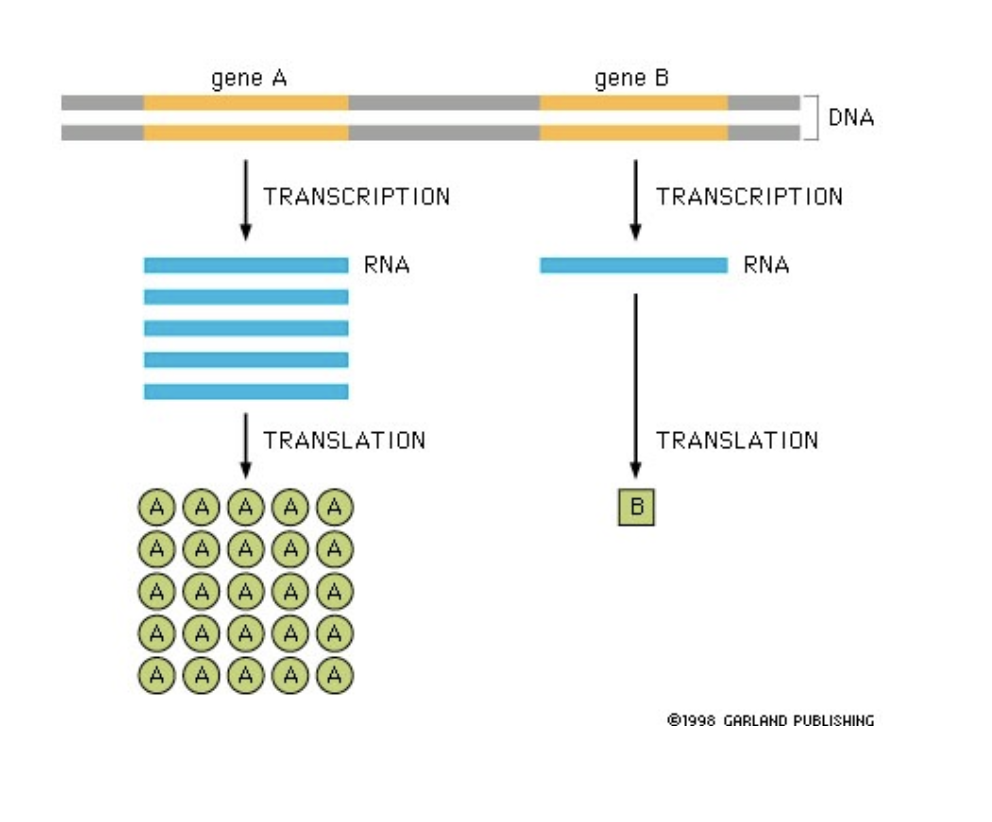
genes
can have intrinsic differences in sequence that cause them to be expressed with different efficiencies
gene expression
can be regulated at any step from transcription to protein activity
transcriptional regulation is most common way of regulating genes, in part because it saves the cell the most energy/resources
often controlled by the presence of small molecules that provide info about the environment, often nutrient availability
the small molecules control transcription by allosteric interaction w/ factors
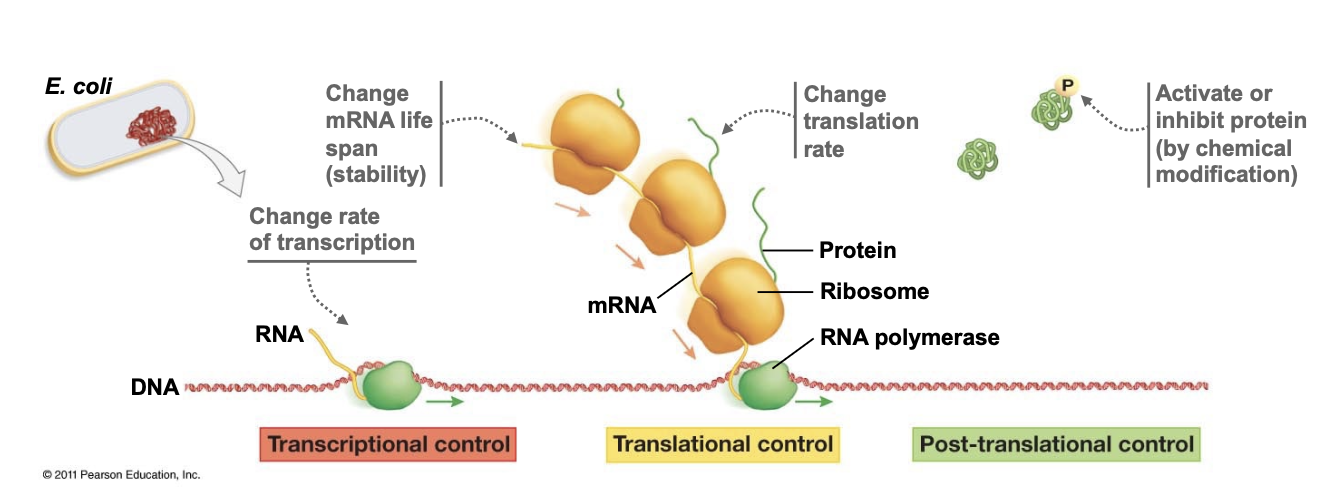
transcription factor
a protein, usually with DNA-binding activity, that influences the rate of transcription of a gene, either positively or negatively
have to find their binding sites in a vast genome of sequence
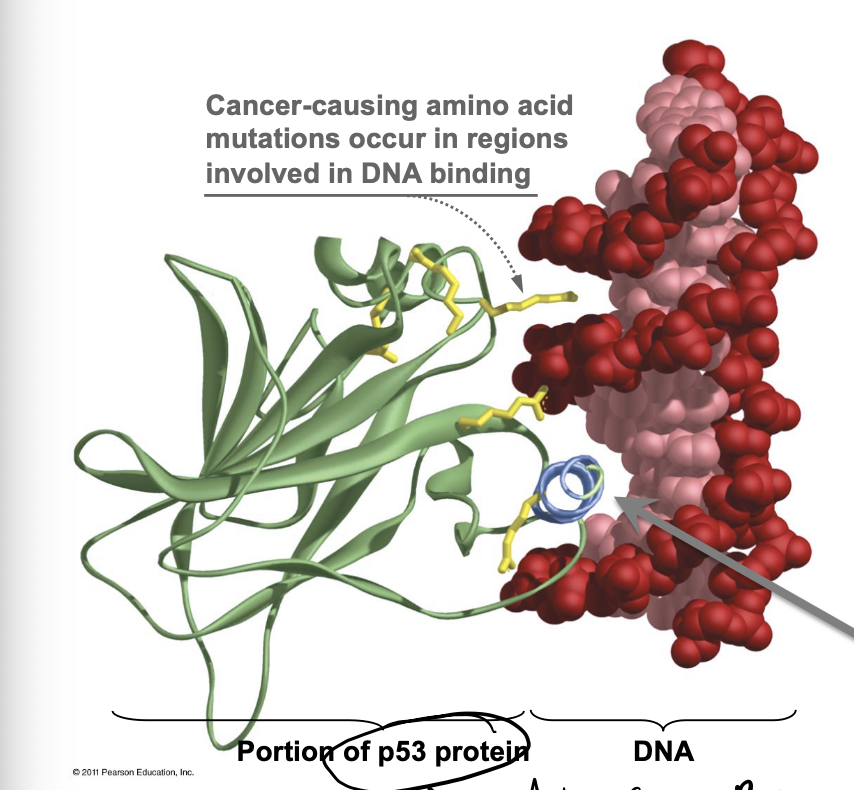
p53 protein
Anti-cancer proteins that we have, prevent tumor formulation
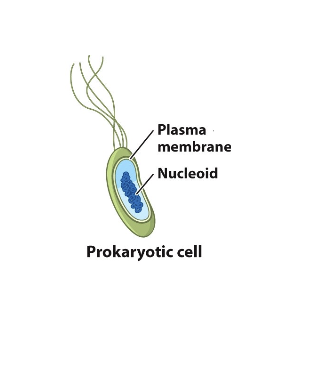
gene regulation
simpler than in eukaryotes because:
DNA is not packaged into chromatin
prokaryotes streamline their processes for efficiency (to minimize energy expenditure
microbiome
all of the microorganisms that live on your skin, in/on your mucosa (ear, mouth, nose), and in your gut
commensal bacteria
“helpful bacteria” that assist with digestion and protect against opportunistic pathogens
prokaryotes and transcriptional regulation
metabolic machines, gene expression is often controlled by the presence of small molecules
regulatory transcription factors
direct regulation by nutrients & small molecules
existence of operons
limited combinatorial control
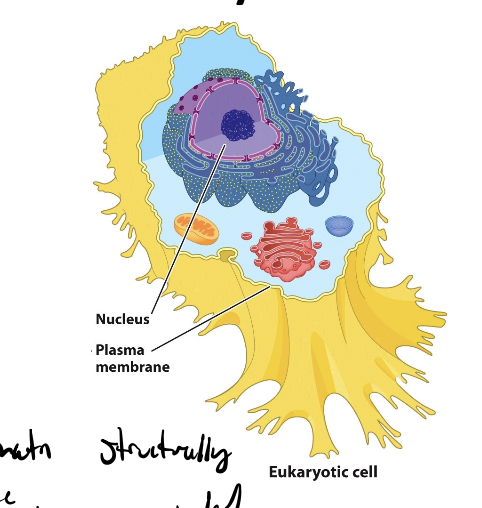
eukaryotes and transcriptional regulation
regulatory transcription factors
regulation by covalent modification of chromatin & its folding state
lots of combinatorial control
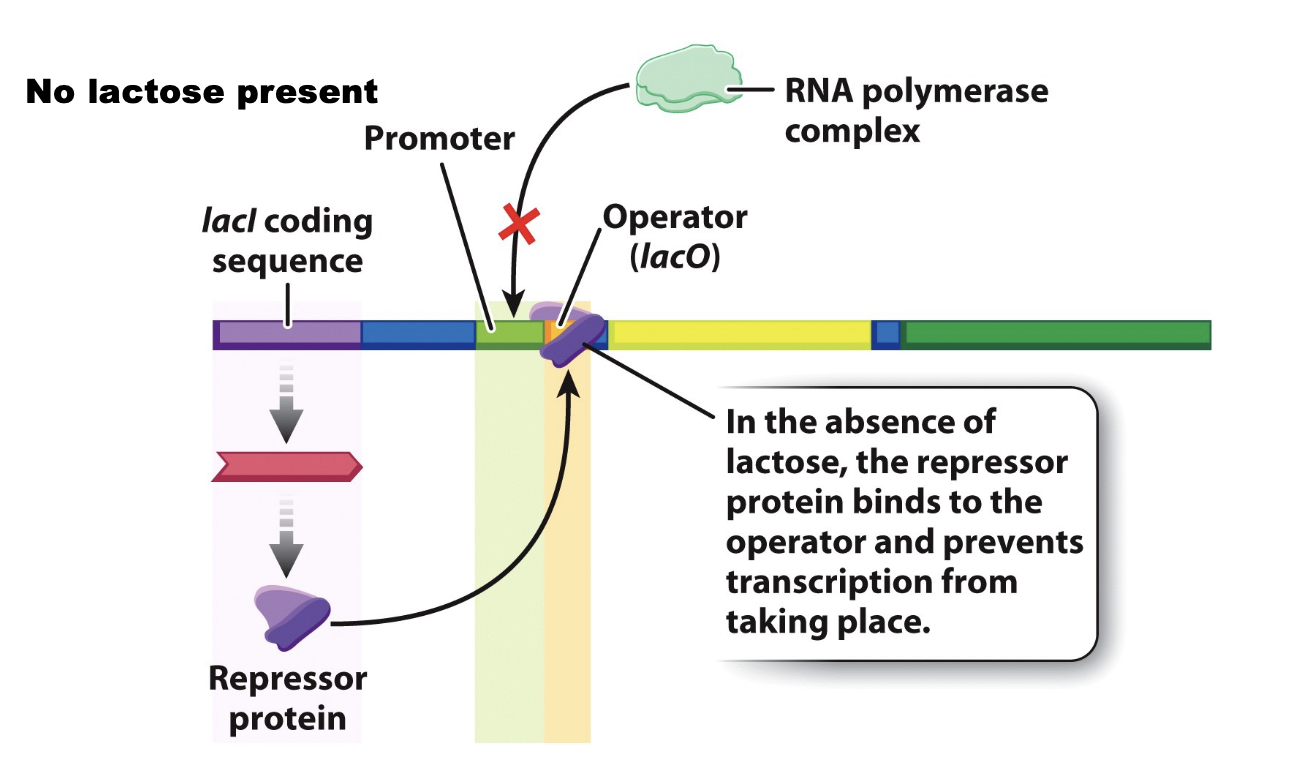
lac operon
illustrates prokaryotic transcription regulation
contains gene coding sequences
lacZ
lacY
lacl
operon
a region of DNA that includes the coding sequence for multiple genes that get transcribed together into a single mRNA (polycistronic mRNA).
common in prokaryotic, very rare in eukaryotes
polycistronic
single molecule of messenger RNA (mRNA) that contains the coding sequences for two or more different proteins
lacl gene
located near the operon and encodes the lac repressor protein
expressed constitutively at a low level
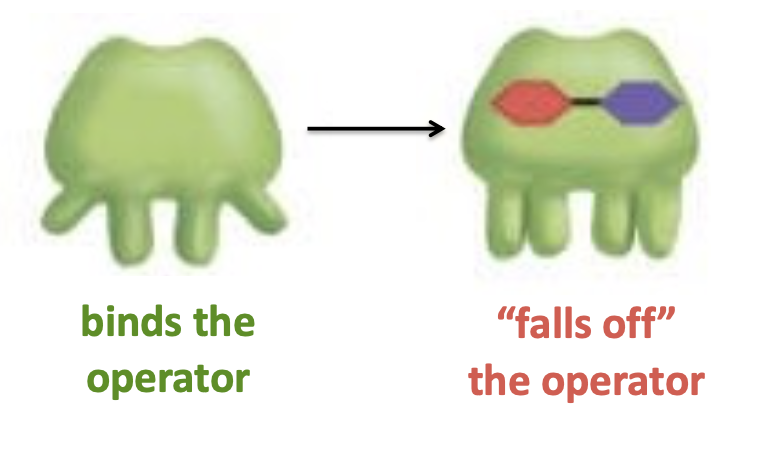
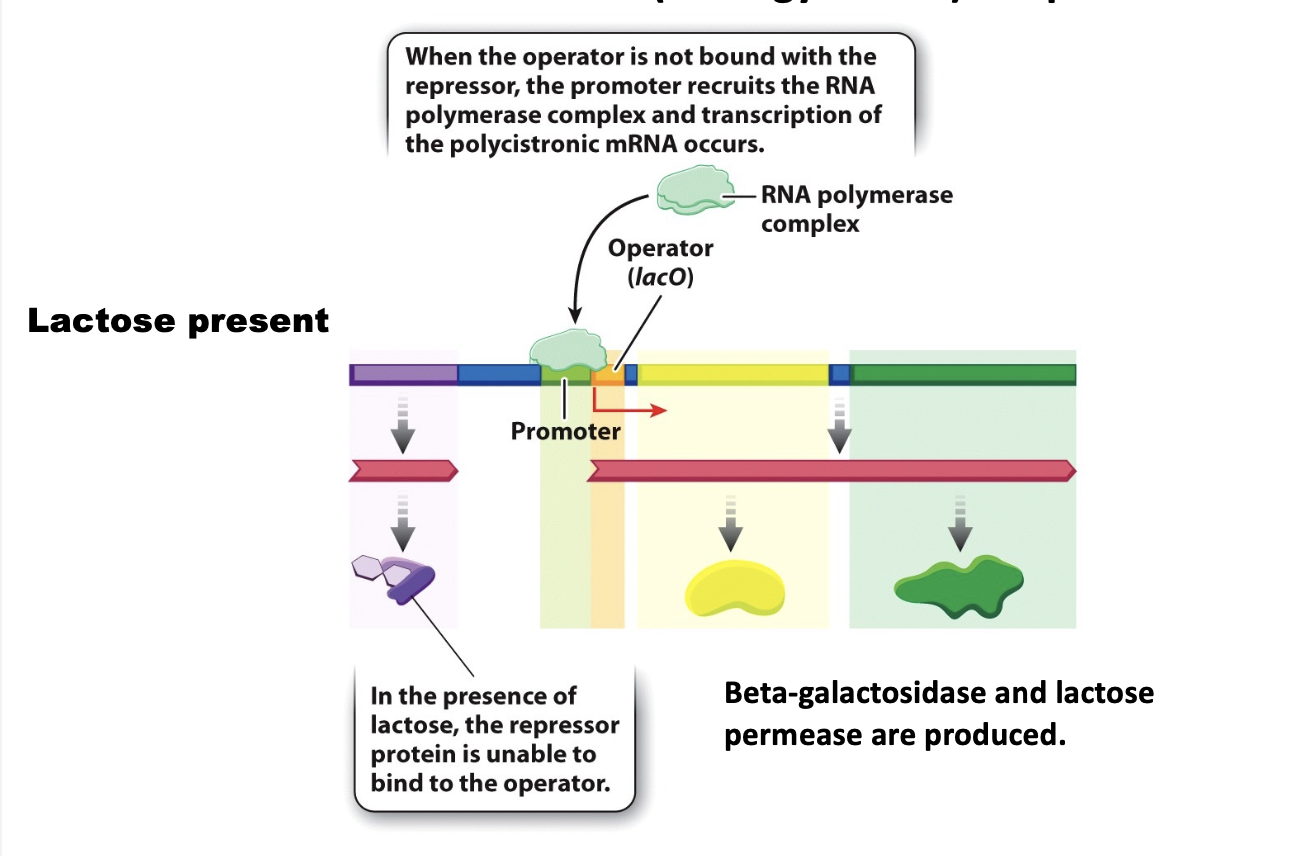
in the absence of lactose, the repressor binds the operator, and consequently RNA pol is not recruited to the promoter
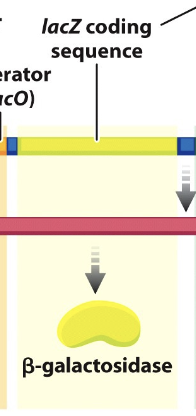
lacZ
the gene for beta-galactosidase, which cleaves the lactose molecule into its glucose and galactose constitutents
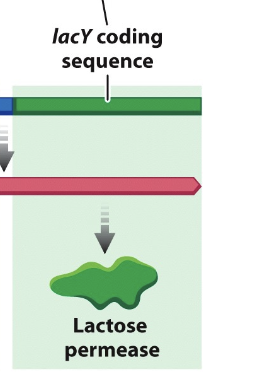
lacY
the gene for lactose permease, which transports lactose from the external medium into the cell
regulatory sequences
promoter
operator — the binding site for a repressor protein
CRP-cAMP binding site —binding site for a positive regulator
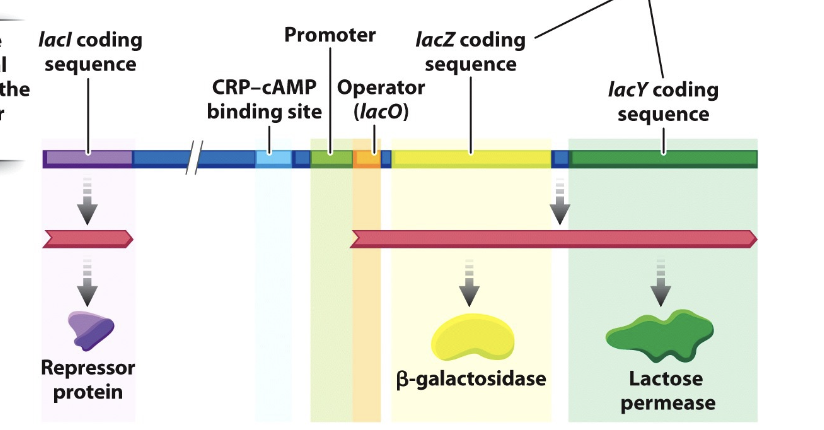
lacI gene repressor
in the presence of lactose, the repressor is allosterically inactivated by binding to allolactose
a conformational change inactivates this through allosteric regulation
allolactose
a lactose metabolite
lacO gene operator
in the presence of lactose, it cannot bind to the operator and transcription is permitted. thus proteins that are needed to use lactose as a carbon source (energy source) are produced.
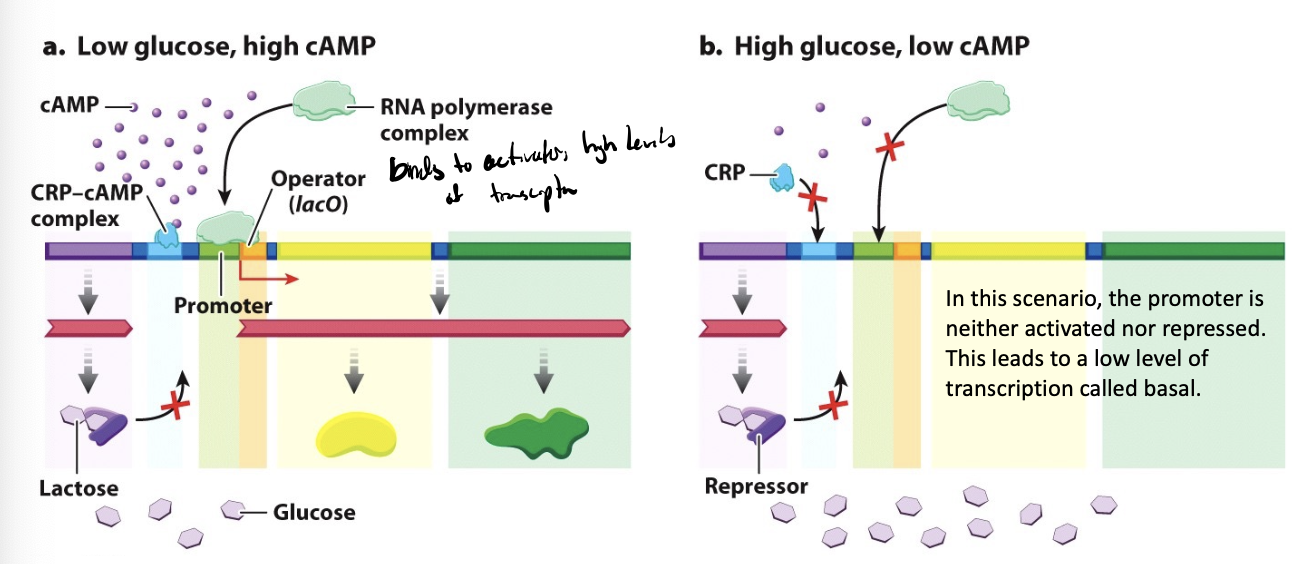
CRP-cAMP complex
a positive regulator of the lac operon that controls transcription based on glucose availability
low glucose → high cAMP
high glucose → low cAMP
provides an additional level of control over transcription that is sensitive to glucose levels
because carbon/energy courses are preferable to others, so if given high glucose and lactose, the cell will use glucose, but if glucose is depleted, it can invest in lactose utilization
lacZ and lacY
loss of a function mutation in the sequence encoding the lac repressor or in the operator sequence causes LacZ and LacY to be constitutively expressed
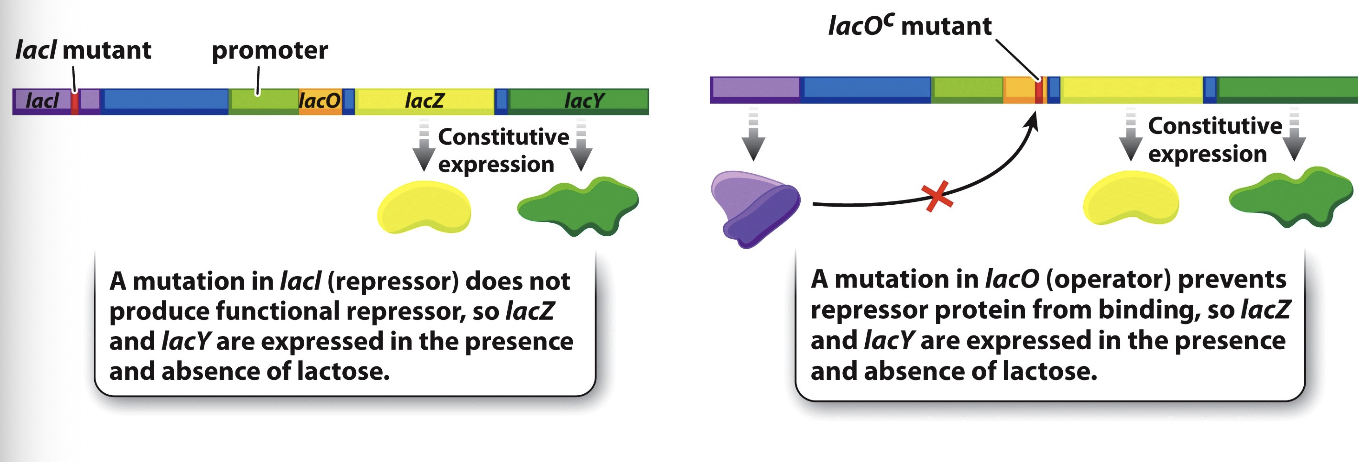
constitutively
under all conditions - regardless of the presence/absence of lactose and glucose
trp operon
encodes genes required for the synthesis of tryptophan, not its utilization; thus, tryptophan makes the repressor bind rather than fall off the DNA
when tryptophan is present, the trp repressor binds the operator, and RNA synthesis is blocked.
when tryptophan is absent, the repressor dissociates from the operation and RNA synthesis proceeds
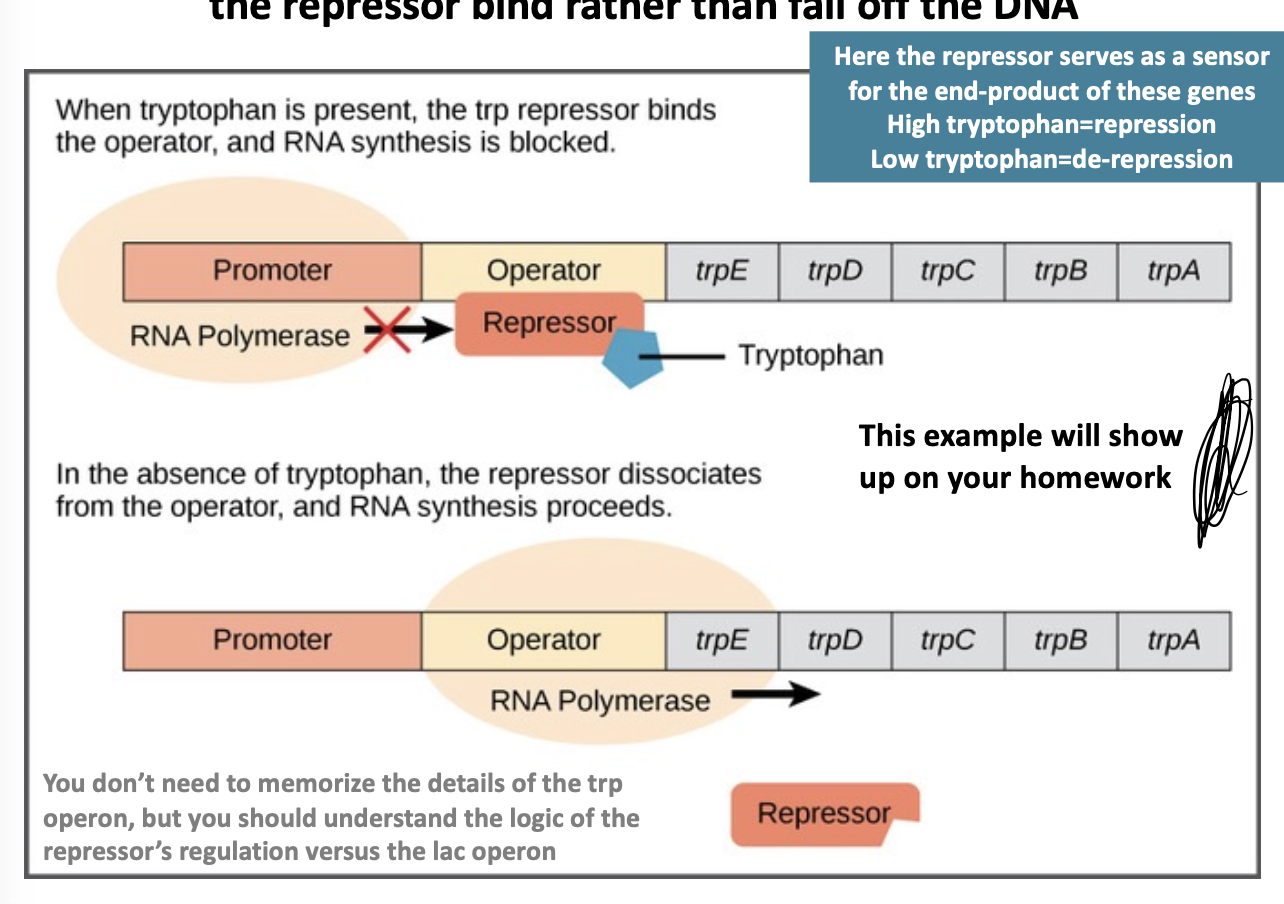
high tryptophan
repression
low tryptophan
de-repression
amino acid bonds
peptide (covalent) bonds