Chapter 19: Variable Number Tandem Repeat Profiling
19.1: Restriction Fragment Length Polymorphism (RFLP)
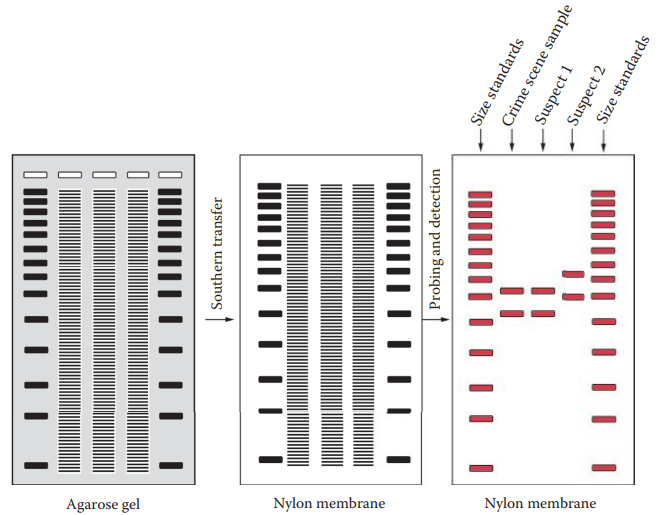
Restriction Fragment Length Polymorphism — the first historical method used in forensic DNA testing.
It utilizes restriction endonucleases that recognize and cleave specific sites along the DNA sequence.
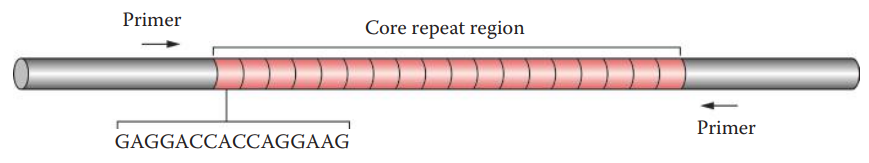
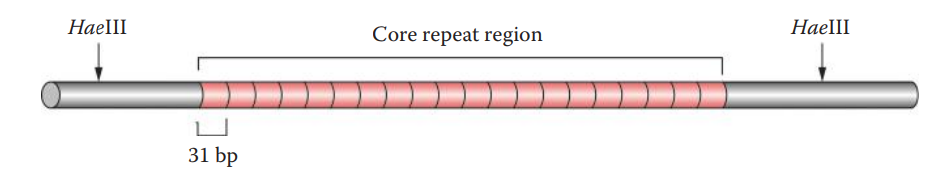
Appropriate restriction endonucleases should be selected so that the genomic DNA is cleaved at sites that flank the VNTR core repeat region.
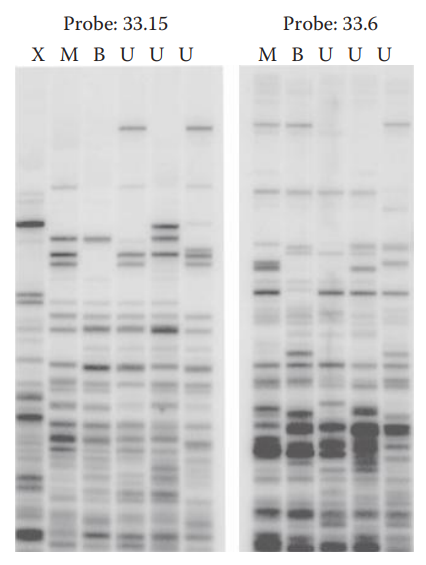
Southern transfer and hybridization technique:
- The DNA is denatured and transferred from the gel to a supporting matrix such as a nylon or nitrocellulose membrane.
- The DNA immobilized on the membrane is then hybridized with a labeled probe.
- Only bands of DNA that have complementary sequences to the probe are recognized by detection systems such as autoradiography.
Common VNTR Loci
| Locus | Chromosome Location | Repeat Unit Length (bp) | Hae III Fragment Size (kb) | Probe |
|---|---|---|---|---|
| D1S7 | 1 | 9 | 0.5-12 | MS1 |
| D2S44 | 2 | 31 | 0.7-8.5 | yNH24 |
| D4S139 | 4 | 31 | 2-12 | pH30 |
| D10S28 | 10 | 33 | 0.4-10 | pTBQ7 |
| D14S13 | 14 | 15 | 0.7-12 | pCMM101 |
| D16S85 | 16 | 17 | 0.2-5 | 3′HVR |
| D17S26 | 17 | 18 | 0.7-11 | pEFD52 |
| D17S79 | 17 | 38 | 0.5-3 | V1 |
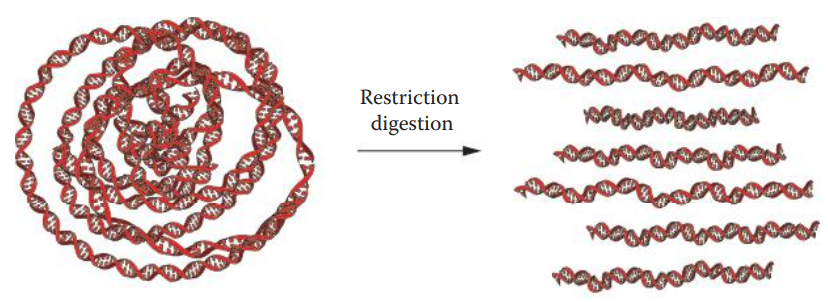
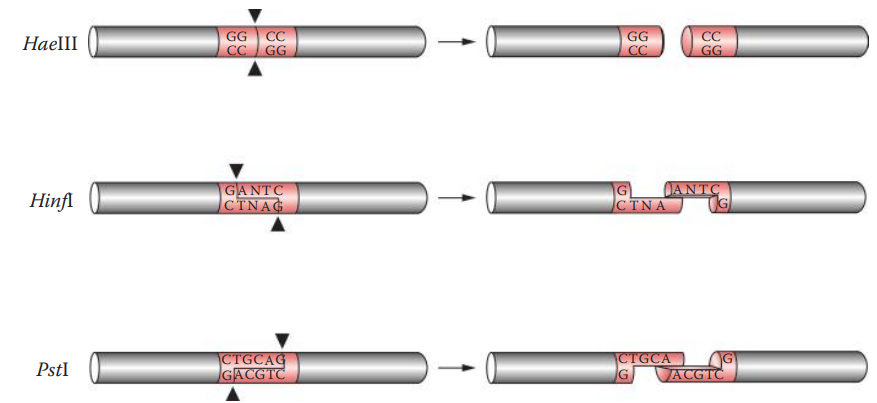
Restriction Endonuclease Digestion
Restriction endonucleases: These are enzymes that cleave the phosphodiester bond of DNA at or near specific recognition nucleotide sequences known as restriction sites.
A restriction site usually is a short motif that is 4–8 bp in length.
It often has a specific palindromic recognition sequence, that is, a segment of double-stranded DNA in which the nucleotide’s sequence is identical to an inverted sequence in the complementary strand.
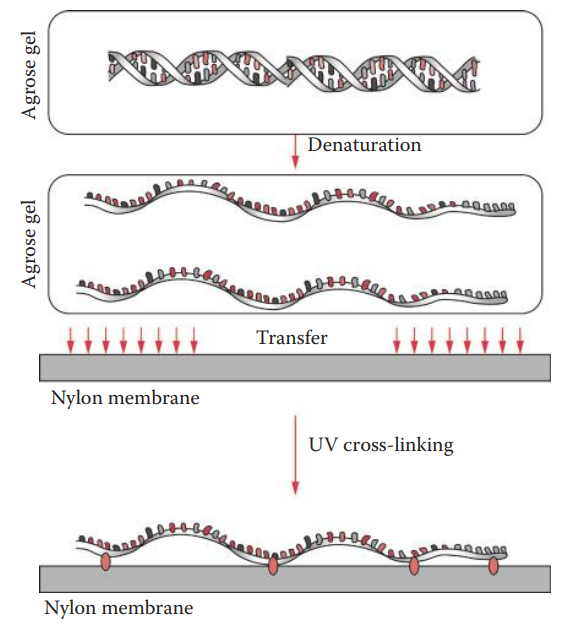
Southern Transfer
It is also known as Southern blotting.
This method can be used to transfer DNA from an agarose gel to a solid matrix so that it can be detected with a hybridization probe.
This technique was named after Sir Edwin Southern.
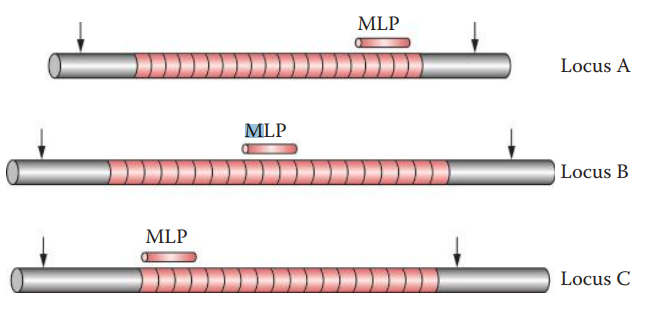
Hybridization with Probes
Multilocus Probe Technique (MLP): It can detect multiple VNTR loci simultaneously.
- It was pioneered by Sir Alec Jeffreys in 1984 and later called DNA Fingerprinting.
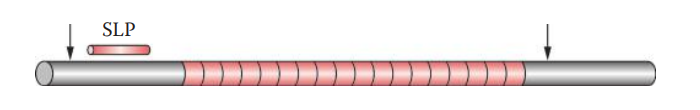
Single-Locus Probe Technique (SLP): It generates a simple pattern called a DNA profile, consisting of one band for a homozygote and two bands for a heterozygote per locus.
Detection
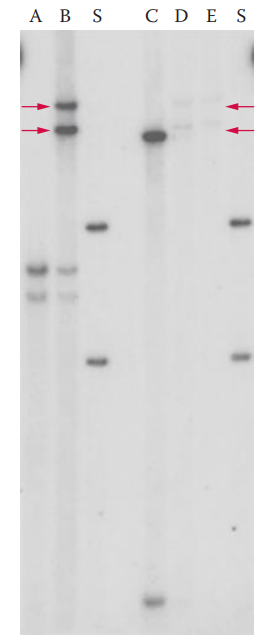
- To detect VNTR loci, a labeled SLP probe is hybridized to the target sequence of DNA, which has been immobilized on a solid matrix such as a piece of nylon membrane transfer.
- Bin: A range of DNA fragments that differ by only a few repeat units.
Factors Affecting RFLP Results
DNA Degradation
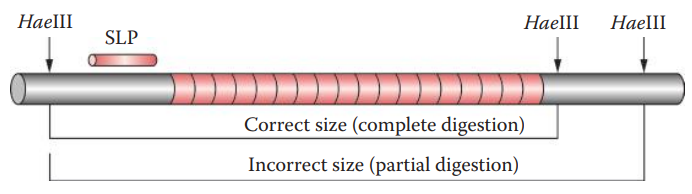
Restriction Digestion–Related Artifacts
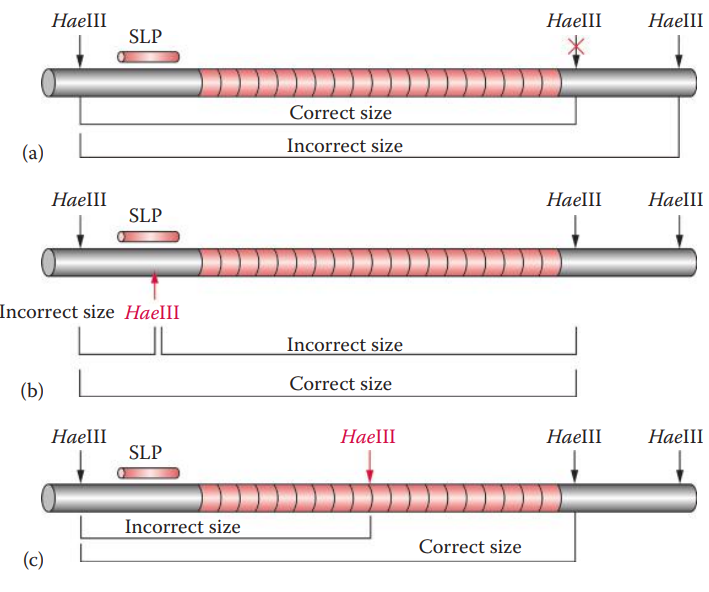
- Partial Restriction Digestion
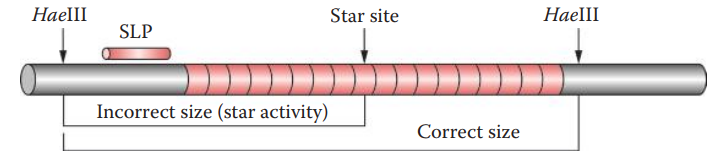
- Star Activity: A deviation of the specificity of a cleavage site of a restriction endonuclease.
- Point Mutations: Caused by the substitution, deletion, or insertion of a single nucleotide.
Electrophoresis and Blotting Artifacts
- Partial Stripping
- Separation Resolution Limits and Band Shifting
- Bands Running off Gel
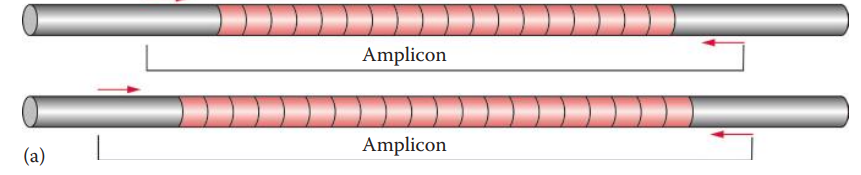
19.2: Amplified Fragment Length Polymorphism (AFLP)
Amplified Fragment Length Polymorphism (AFLP): A PCR-based technique that uses selective amplification of a subset of digested DNA fragments to generate and compare unique fingerprints for genomes of interest.
D1S80: Locus used by forensic DNA laboratories for AFLP analysis.
D1S80 loci are detected as discrete alleles and thus can be compared directly to an allelic ladder on the same gel.
The AFLP technique requires less DNA than the RFLP method and performs better for degraded samples. The AFLP method at the D1S80 locus can be analyzed in a multiplex fashion with an amelogenin locus.
The amelogenin gene is used for forensic sex-typing applications. Typing the amelogenin gene enables the determination of the sex of the contributor of a biological sample.