AP Biology Unit 6 Gene Expression and Regulation
1/65
Earn XP
Description and Tags
Vocab from Unit 6 of AP Biology Gene Expression and Regulation
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
66 Terms
Nucleotide
This is the basic building block of nucleic acids (RNA and DNA). It consists of a sugar molecule (either ribose in RNA or deoxyribose in DNA) attached to a phosphate group and a nitrogen-containing base.
The bases in DNA are: Adenine, bonding to Thymine; and Guanine, bonding to cytosine
The bases in RNA are: same as DNA but Uracil takes the place of thymine.
Purine
This a nitrogenous base with a double ring structure, consisting of a six-membered ring fused with a five-membered ring. In other words, it has 2 nitrogen-containing rings. They are larger then pyrimidines
Examples: Adenine and Guanine
Pyrimidines
These are nitrogenous bases with a single six-membered ring structure. These are smaller than purines.
Examples: Cytosine, Thymine, and Uracil
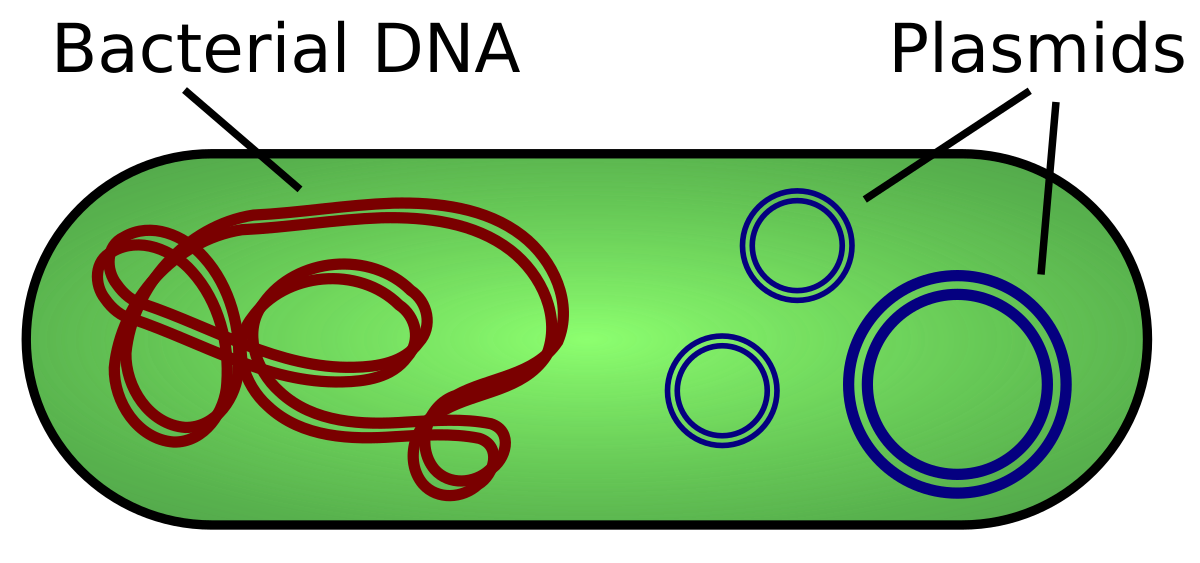
Plasmids
These are small, circular, double-stranded DNA molecule that is distinct from a cell's chromosomal DNA. They naturally exist in bacterial cells, and they also occur in some eukaryotes. Often, the genes carried here provide bacteria with genetic advantages, such as antibiotic resistance.
They can be found in nucleus of eukaryotes or the cytosol of prokaryotes.
Polypeptide
This is another word for a strand of amino acids. Although many proteins consist of a single BLANK, some are made up of multiple. Genes that specify BLANKs are called protein-coding genes.
DNA Polymerase
This synthesizes DNA by adding nucleotides one by one to a growing chain. The enzyme always needs a template and adds on to the 3’ end.
The enzyme even proofreads the work to see if it is correct, removing most of the incorrect nucleotides.
DNA Polymerase I
Removes RNA primers and replaces them with DNA
Fills gaps in DNA that occur during replication, repair, and recombination
DNA Polymerase II
Proofreads and edits DNA, mainly in the lagging strand
Repairs damaged DNA strands
Catalyzes the repair of nucleotide base pairs
DNA Polymerase III
The main enzyme responsible for replicating DNA in bacterial cells
Extends primers to make the bulk of the new DNA
DNA Ligase
It joins DNA fragments on the lagging strand of DNA.
DNA Helicase
This catalyzes the disruption of the hydrogen bonds that hold the two strands of double-stranded DNA together.
aka it opens up the DNA at the replication fork.
DNA Primase
This primes DNA synthesis (gets it started). It makes an RNA primer, or short stretch of nucleic acid complementary to the template, that provides a 3' end for DNA polymerase to work on. A typical primer is about five to ten nucleotides long.
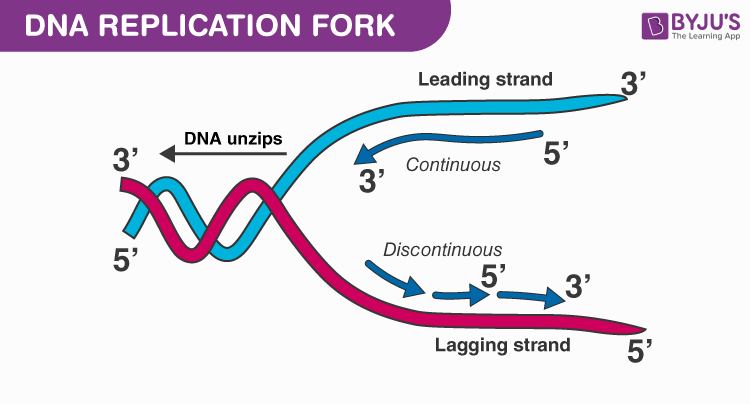
Leading Strand
This is the strand that runs in the 5' to 3' direction in the replication fork. This strand is continuously synthesized.
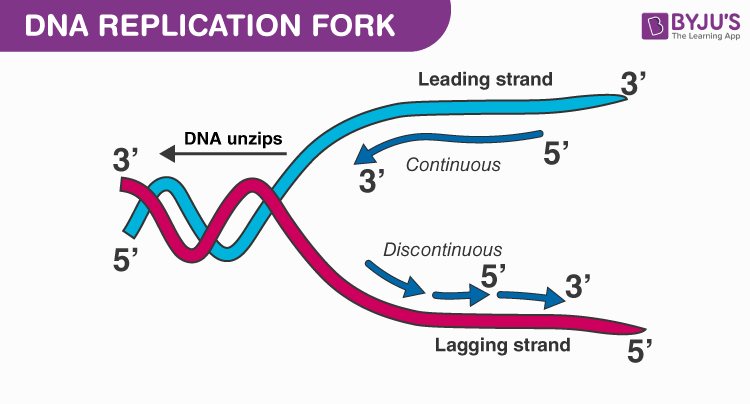
Lagging Strand
Is one of two strands of DNA found at the replication fork, or junction. This strand requires a slight delay before undergoing replication, and it must undergo replication discontinuously in small fragments (Okazaki Segments).
Sliding Clamp
This is a protein that helps out in DNA replication and holds DNA polymerase III molecules in place as they synthesize DNA. It is a ring-shaped protein and keeps the DNA polymerase of the lagging strand from floating off when it re-starts at a new Okazaki fragment.
Topoisomerase
This is an enzyme that assists in DNA replication. This enzyme prevents the DNA double helix ahead of the replication fork from getting too tightly wound as the DNA is opened up. It acts by making temporary nicks in the helix to release the tension, then sealing the nicks to avoid permanent damage.
Basically, it relaxes the supercoil at the replication fork.
Gene Expression
the process by which the information encoded in a gene is turned into a function. This mostly occurs via the transcription of RNA molecules that code for proteins or non-coding RNA molecules that serve other functions.
Transcription
This is the first step in gene expression. The goal is to make a RNA copy of a gene's DNA sequence. For a protein-coding gene, the RNA copy, or transcript, carries the information needed to build a polypeptide (protein or protein subunit).
This is performed by RNA polymerases and occurs in 3 steps, initiation, elongation, and termination.
RNA Polymerase
This is the enzyme responsible for copying a DNA sequence into an RNA molecule by adding complementary nucleotides to the growing RNA strand, essentially acting as the main catalyst for the process of transcription.
Specifcally, it builds an RNA strand in the 5’ to 3’ direction, adding each nucleotide to the 3’ end.
mRNA
it acts as a messenger to the cell's machinery that makes proteins, giving instructions for what proteins to make and how to make them.
It carries the copy of the gene to the ribosomes in transcription.
rRNA
This is a non-coding RNA that forms a major part of ribosomes, essential for protein synthesis in all cells.
Transfer RNA
These are molecular "bridges" that connect mRNA codons to the amino acids they encode. One end of each BLANK has a sequence of three nucleotides called an anticodon, which can bind to specific mRNA codons. The other end of the BLANK carries the amino acid specified by the codons.
This type of RNA reads mRNA .
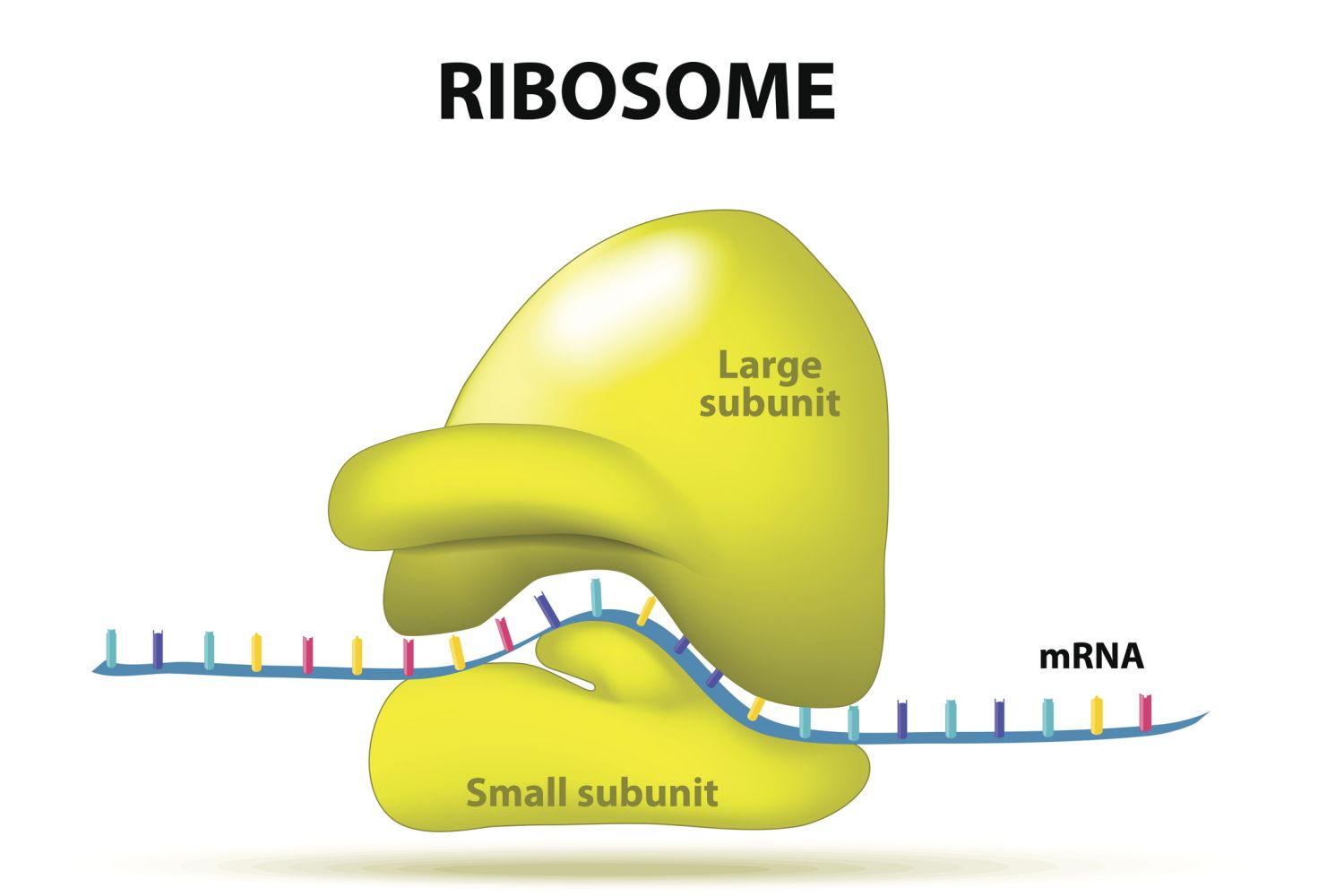
Ribosomes
These are the structures where polypeptides (proteins) are built. They are made up of protein and RNA (ribosomal RNA, or rRNA). Each one has two subunits, a large one and a small one, which come together around an mRNA.
They provide a set of handy slots where tRNAs can find their matching codons on the mRNA template and deliver their amino acids. These slots are called the A, P, and E sites. Not only that, but the BLANK also acts as an enzyme, catalyzing the chemical reaction that links amino acids together to make a chain.
Initiation
The ribosome assembles around the mRNA to be read and the first tRNA (carrying the amino acid methionine, which matches the start codon, AUG). This setup, called the initiation complex, is needed in order for translation to get started. is the first step of transcription.
RNA polymerase binds to a sequence of DNA called the promoter, found near the beginning of a gene. Each gene (or group of co-transcribed genes, in bacteria) has its own promoter. Once bound, RNA polymerase separates the DNA strands, providing the single-stranded template needed for transcription.
Elongation
This is the second step of transcription. One strand of DNA, the template strand, acts as a template for RNA polymerase. As it "reads" this template one base at a time, the polymerase builds an RNA molecule out of complementary nucleotides, making a chain that grows from 5' to 3'. The RNA transcript carries the same information as the non-template (coding) strand of DNA, but it contains the base uracil (U) instead of thymine (T).
Termination
This is the third step of transcription. These are sequences that signal that the RNA transcript is complete. Once they are transcribed, they cause the transcript to be released from the RNA polymerase.
5’ Cap
This is a modified guanine nucleotide that plays a crucial role in mRNA stability, translation, and nuclear export. It protects the mRNA from exonucleases (degradation), promotes ribosomal binding, and regulates nuclear export.
The ribosome binds to this to initiate translation.
3’ Poly A tail
This helps terminate transcription. It is made up of a bunch of Adenine nucleotides. It also plays a crucial role in mRNA stability, translation, and nuclear export. It protects the mRNA from exonucleases (degradation), promotes ribosomal binding, and regulates nuclear export.
pre-mRNA
In bacteria, RNA transcripts can act as messenger RNAs (mRNAs) right away. In eukaryotes, the transcript of a protein-coding gene is called a BLANK and must go through extra processing before it can direct translation.
Eukaryotic pre-mRNAs must have their ends modified, by addition of a 5' cap (at the beginning) and 3' poly-A tail (at the end).
Many eukaryotic pre-mRNAs undergo splicing. In this process, parts of the pre-mRNA (called introns) are chopped out, and the remaining pieces (called exons) are stuck back together.
Splicing
Parts of the pre-mRNA are chopped out (introns), and the remaining pieces (called exons) are stuck back together. This gives the mRNA its correct sequence. (If the introns are not removed, they'll be translated along with the exons, producing a "gibberish" polypeptide since introns don’t code for amino acids.)
Alternative Splicing
Regular splicing allows for this process, which is when more than one mRNA can be made from the same gene. Through alternative splicing, we (and other eukaryotes) can encode more different proteins than we have genes in our DNA.
Translation
This takes place in ribosomes. The sequence of the mRNA is decoded to specify the amino acid sequence of a polypeptide. The name indicates that the nucleotide sequence of the mRNA sequence must be translated into the completely different "language" of amino acids.
This is the process of using information in an mRNA to build a polypeptide.
Codons
These are a sequence of three DNA or RNA nucleotides that corresponds with a specific amino acid or stop signal during protein synthesis. DNA and RNA molecules are written in a language of four nucleotides; their are 61 codons that specify amino acids.
In translation, these are read and one BLANK is a "start" BLANK that indicates where to start translation. It specifies the amino acid methionine, so most polypeptides begin with this amino acid. Ex. AUG, GUG, UUG, TTG. Three more BLANKs do not specify amino acids. These stop BLANKs, UAA, UAG, and UGA, tell the cell when a polypeptide is complete.
These relationships between BLANKs and amino acids are called the genetic code.
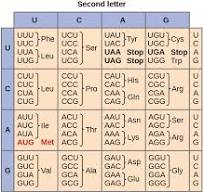
Genetic Code
This is the relationship between codons and amino acids (stop signals). It is often summarized in a table. Many amino acids are represented in the table by more than one codon. For instance, there are six different ways to "write" leucine in the language of mRNA (see if you can find all six).
Central Dogma
This is a theory stating that genetic information flows only in one direction, from DNA, to RNA, to protein, or RNA directly to protein.
Non-coding Strand
This is the strand of DNA that serves as the template for the synthesis of a matching (complementary) RNA strand by an enzyme called RNA polymerase. This RNA strand is the primary transcript.
Primary Transcript
This is the RNA strand produced by the transcription of DNA. It carries the same sequence information as the non-transcribed strand of DNA, sometimes called the coding strand. However, the BLANK and the coding strand of DNA are not identical, thanks to some biochemical differences between DNA and RNA.
Promoter
This is a specific DNA sequence located upstream of a gene that signals the start of transcription, where RNA polymerase and other transcription factors bind to initiate the process of creating RNA from DNA. Strong ones result in high expression, while weak ones lead to lower expression.
In eukaryotes, each gene has its own BLANK, whereas in prokaryotes, its normal for a few genes to be grouped together for one BLANK.
Operon
This is when a cluster of genes are under the control of one promotor in bacteria or prokaryotes (very rare in humans). This feature allows protein synthesis to be controlled coordinately in response to the needs of the cell.
Some are inducible, meaning that they can be turned on by the presence of a particular small molecule. Others are repressible, meaning they are on by default but can be turned off by a particular small molecule called a corepressor.
Regulatory Gene Expression
These are specific DNA or RNA regions that control gene expression, determining when, where, and how much a gene is expressed, ultimately influencing an organism's traits. They control transcription of the operon.
Repressors
These attach to a sequence of DNA (operator) after the promoter and block RNA polymerase from doing transcription.
Activator
These are proteins that increase the rate of transcription of specific genes by binding to DNA sequences, often enhancers or promoter-proximal elements, and interacting with other proteins to facilitate RNA polymerase binding and elongation.
Small molecules called inducers bind to BLANKs to activate them.
Lac Operon
This is an inducible operon that encodes enzymes for metabolism of the sugar lactose. It turns on only when the sugar lactose is present (and other, preferred sugars like glucose are absent). The inducer in this case is allolactose, a modified form of lactose. The inducer in this case is allolactose, a modified form of lactose.
The reason that transcription is less when glucose is present is because glucose is preferred over lactose for transcription since it is a very simple sugar.
Allolactose
This an isomer of lactose and is present when lactose is present to act as an inducer of transcription. It does this by binding to the lac opressor, preventing it from binding to the operator site.
Isomer: compounds that have the same molecular formula but are structurally different
Trp Operon
This is essential for the production of tryptophan (amino acid). It is regulated by a negative feedback loop. It’s part of the E.coli genome.
When there is low tryptophan amounts, RNA polymerase can go ahead and start transcribing and creating more tryptophan.
When there is high amts of tryptophan, the tryptophan acts as a co-repressor and binds to the trp repressor, causing it to bind to the operator and blocking RNA Polymerase from transcribing.
Heterochromatin
This is a type of chromatin that is highly condensed, gene-poor, and transcriptionally inactive. Typically, genes that are located within this are not expressed.
Euchromatin
This is a type of chromatin that is less condensed, gene rich, and transcriptionally active. Typically, genes that are located within this are expressed.
Mutagen
A chemical or physical substance or event that can cause genetic changes.
There are two types:
Endogenous: found inside the organism
Exogeneous: comes from outside the affected organism.
Endogenous Mutagen
These are found inside the organism and an example includes Reactive Oxygen Species (ROS) such as superoxide (oxygen with an extra electron) or hydrogen peroxide (H2O2).
During metabolism, the mitochondria produce lots of ROS, which becomes harmful when there is too much since it reacts with DNA and can damage genetic code.
Examples of genetic code damage include the double strand of DNA breaking from ROS or base modification, where the nucleic acid bases are changed or swapped.
“Oxidative Stress” is when you have too much ROS, but Anti-Oxidants can ensure the ROS doesn’t damage DNA.
Exogenous Mutagen
These come from outside the affected organism. Physical examples include UV and ionizing (X-ray, gamma) radiation,
Chemical examples include:
Intercalating agents: like ethidium bromide (EtBr) and acridine dyes, insert themselves between DNA base pairs, disrupting the DNA structure and causing mutations, often frameshift mutations
Base Analogs: ike 5-chlorouracil, can substitute for normal DNA bases during replication, leading to mismatched base pairing and mutation
Carcinogens
These CAN be mutations but not necessarily. They can lead to cancer and cause tumors.
ex. tobacco, asbestos (group of minerals that were used in construction), UV radiation
Aneuploidy
This is a genetic mutation in which there are extra or missing chromosomes.
Triploidy
This is having 3 copies of a particular chromosome. In animals in can lead to sterility.
Polyploidy
This is having multiple sets of homologous chromosomes. It can cause increased vigor in plants (rowing faster and bigger), but also sterility.
Recombination
This is the exchange of genetic material between different viral genomes, often occurring when multiple viruses infect the same host cell, leading to the creation of new viral strains with combined genetic characteristics
Retrovirus
This is a specific type of virus that uses RNA as its genomic material. It creates a DNA copy of its RNA genome using an enzyme called Reverse Transcriptase and inserts the DNA copy into a host cell that it invades. The viral DNA will be transcribed and translated, thus changing the genome of that cell.
Horizontal Acquisition
This refers to the transfer of genetic material between organisms that are not parent and offspring
Transformation
A bacterium takes up a piece of DNA floating in its environment.
Transduction
DNA is accidentally moved from one bacterium to another by a virus.
Conjugation
DNA is transferred between bacteria through a tube between cells. In most cases, the DNA is in the form of a plasmid.
Transposable Elements
These are chunks of DNA that “jump” from one place to another. They can move bacterial genes that give bacteria antibiotic resistance or make them disease-causing.
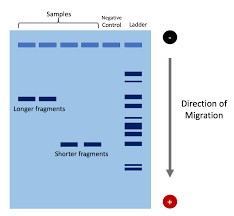
Gel Electrophoresis
This is a laboratory technique used to separate DNA, RNA, or protein fragments based on their size and charge. It does this by applying an electric field to a gel matrix, creating a positive and negative ends. The DNA, being negatively charged, goes to the positive end, separating fragments by size since smaller molecules faster and further than larger ones.
The band patterns can also be used to identify people.
Polymerase Chain Reaction
This is a laboratory technique used to rapidly amplify specific DNA sequences, creating millions of copies from a small sample, enabling the detection and analysis of genetic material.
It does this by:
1. Denaturing the DNA (heat to 96C)
2. Primers are added after DNA is cooled to 55C (annealing)
3. DNA is replicated
4. Process is repeated desired amount (25 - 35 times)
DNA Sequencing
This is a laboratory technique used to determine the precise order of nucleotides (A, T, C, and G) in a DNA molecule.
Some techniques include:
labeling the nucleotides with a fluorescent dye to read and build copies of DNA
Bacterial Transformation
Additional genes are added to bacterial genomes in order to study their function.
Aka “molecular cloning”
Can be used for biopharmaceuticals, gene therapy, and gene analysis.
Splice out DNA segments that you want to use using a restriction enzyme
Genes are inserted into bacterial plasmid and combined with ligase to make a recombinant plasmid.
Plasmid are introduced to bacteria through transformation
Dimers
These are formed when two adjacent pyrimidine bases (thymine or cytosine) are covalently linked by a cyclobutane ring, typically induced by ultraviolet (UV) light exposure, causing DNA damage and hindering replication and transcription