BioImaging Subjects 1 & 2
1/60
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
61 Terms
Signals
math functions of 1+ ID variables, model physical processes
Image (2D)
function of 2 ID real valued variables (pixels)
visual representation of 2D signal
Delta/Impulse Function
models a point source (char resolution)
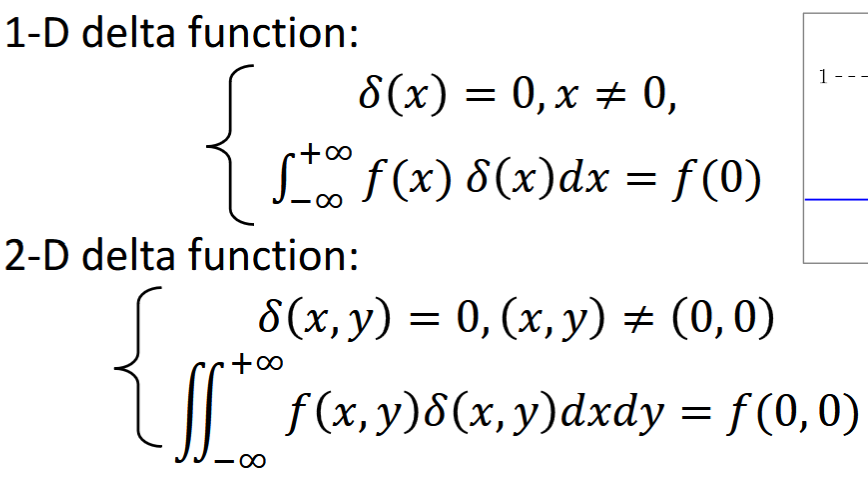
Delta Function Properties
Shift, Scale, Even
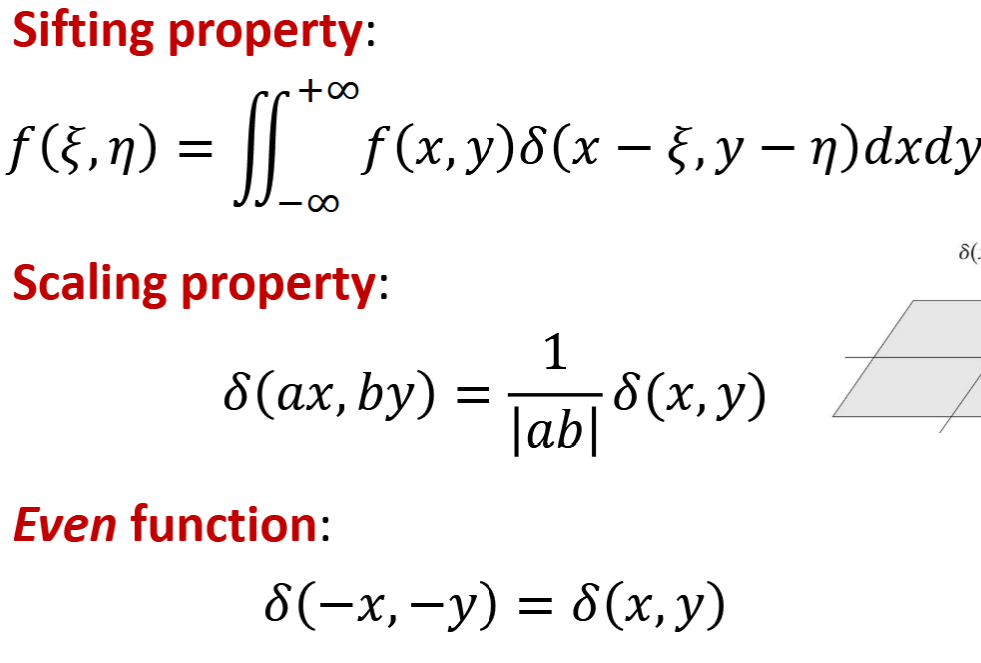
Comb Function
shows pixel grid
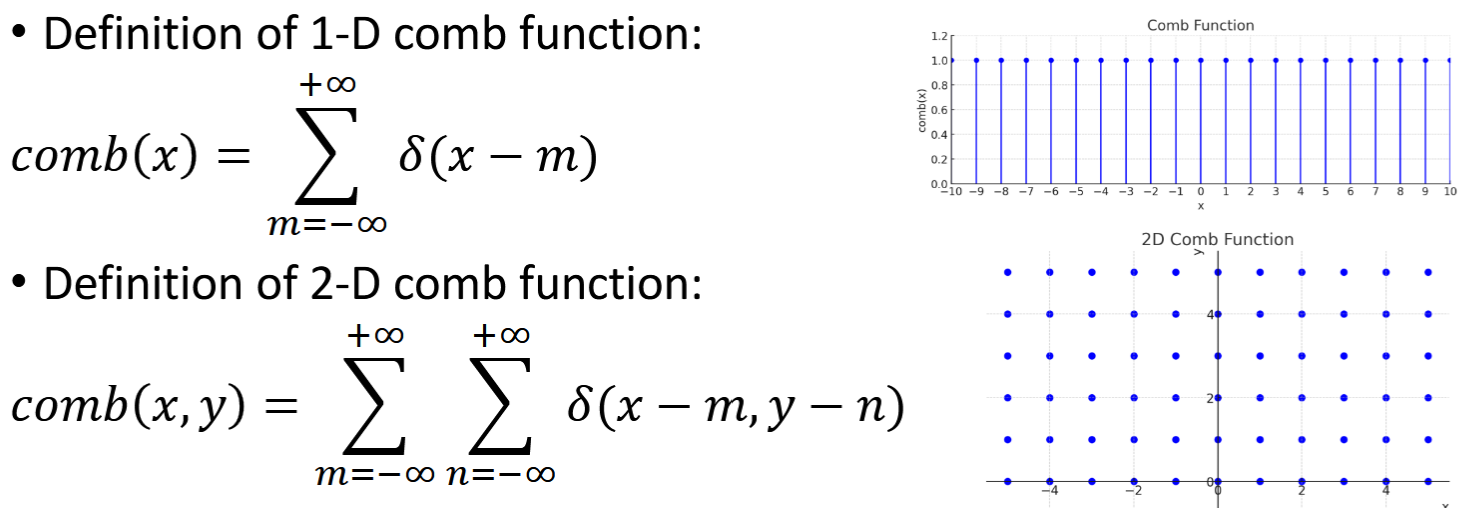
Sampling
converts continuous signals to discrete signals. Summations
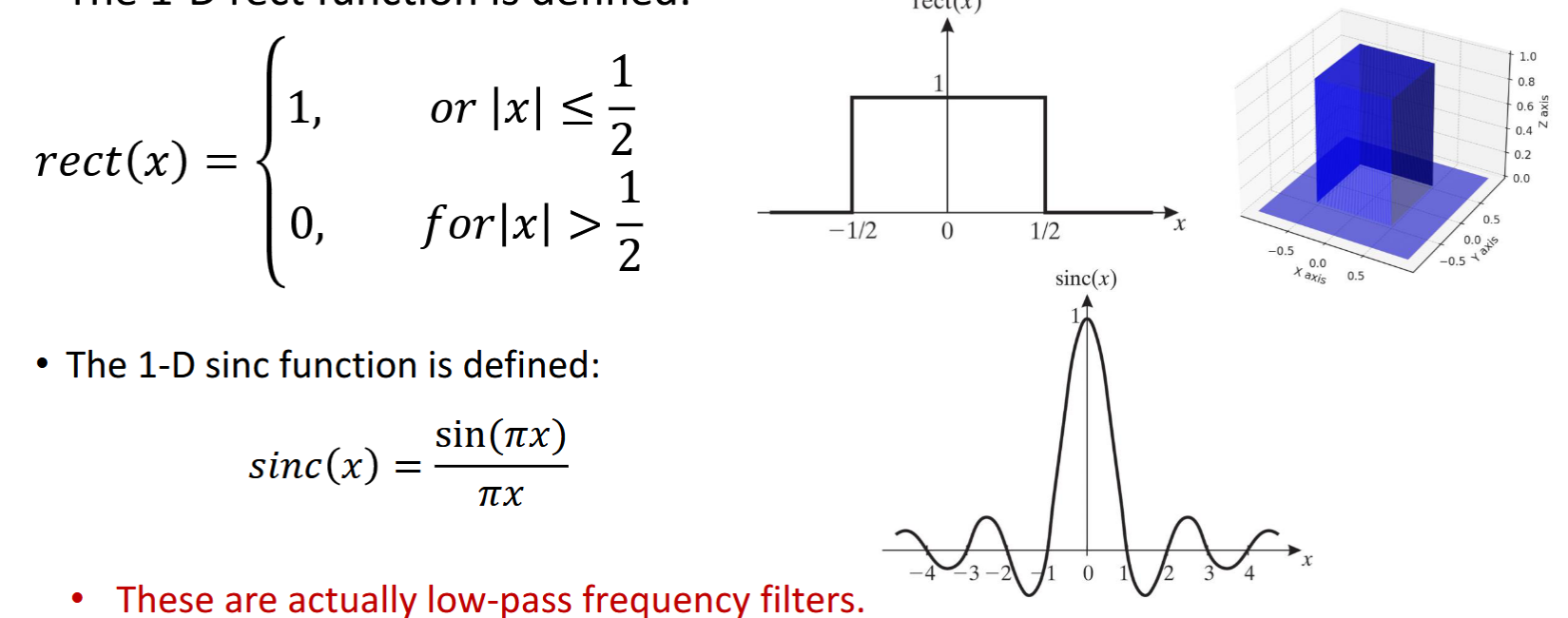
Rect and Sinc Functions
Low pass filters, fourier transforms of each other
Separable Signals
most medical images, 2D functions that can be reduced to 1D operations. f(x,y)=f(x)*f(y)
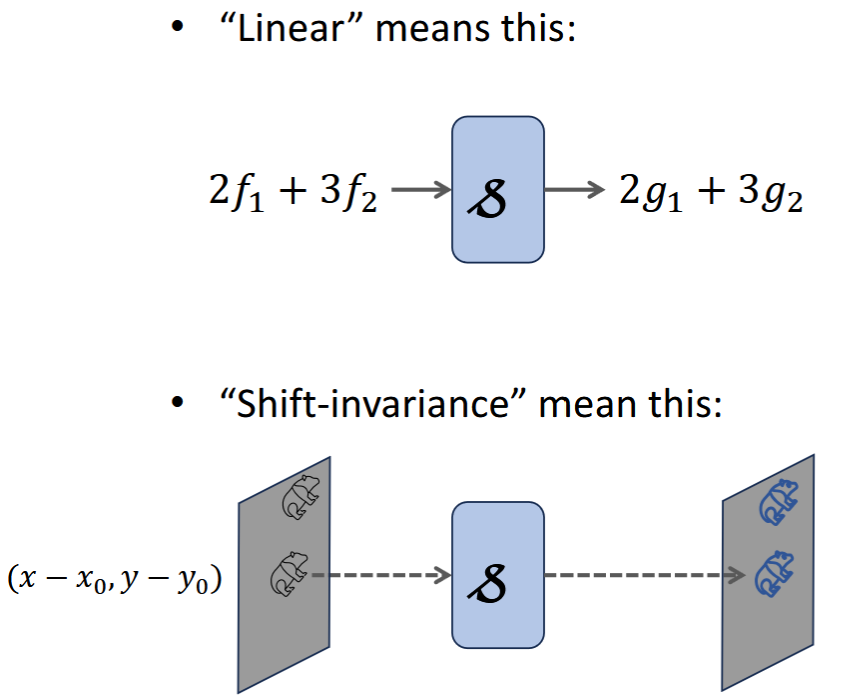
System
transformation 𝓈 of input signal f to output signal g.
𝓈 = characterization of the system, blurring function
Linear System
Scaleable - input x C = output x C
Superposition - 2 inputs => output = output1+output2
most medical systems
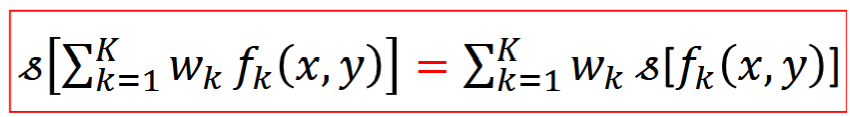
Shift Invariant System
translation of input => same translation of output
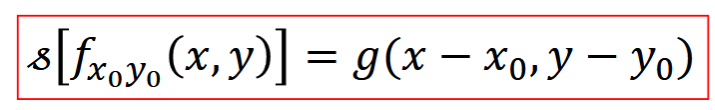
LSI System
Linear and shift invariant system, most medical
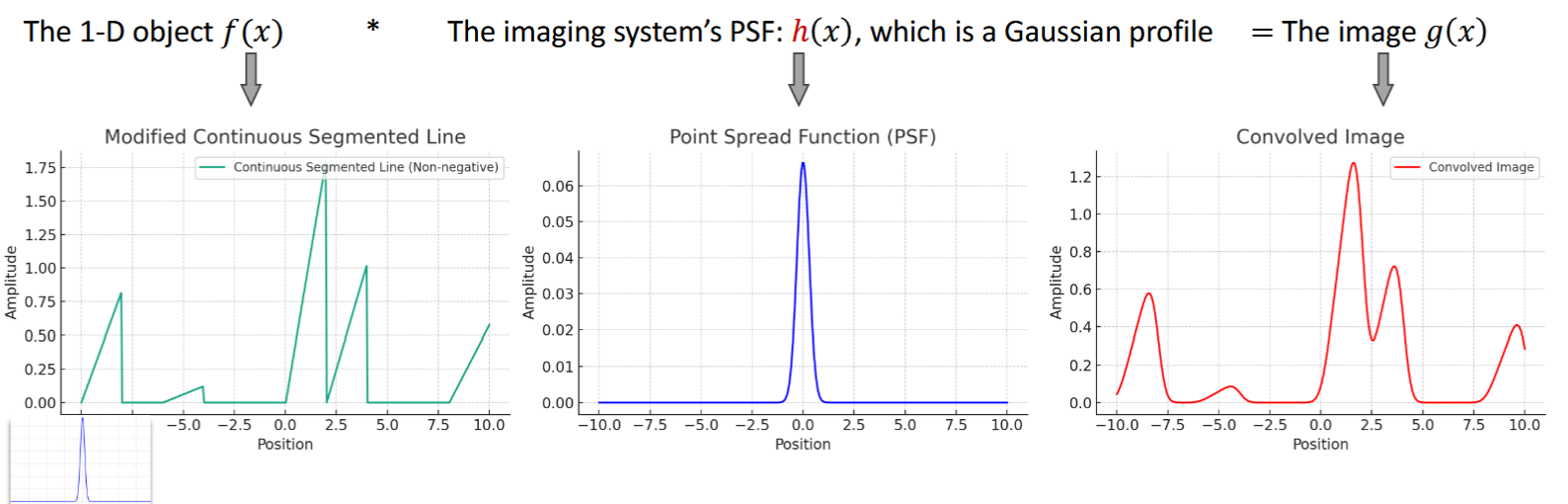
Point Spread Function (PSF)
output of system to a delta function, h, superposition integral, defines system, mostly gaussian profile
for LSI: g(x,y)=h(x,y)*f(x,y)
*= convolution operator
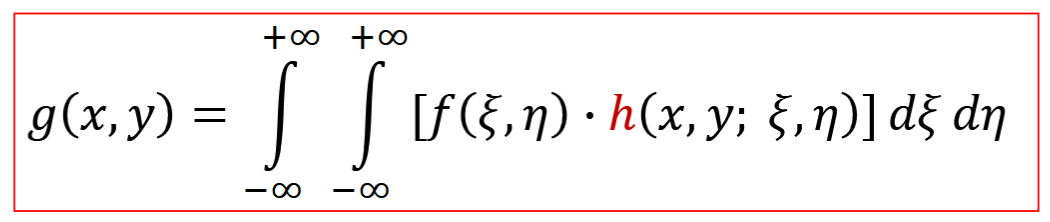
LSI System Connections
Cascade: h=h1*h2
Parallel: h=h1+h2
Associativity: h2*[h1*f]=h1*[h2*f]=[h1*h2]*f
Commutativity: h1*h2=h2*h1
Distributivity: g(x,y)=h1*f+h2*f=(h1+h2)*f
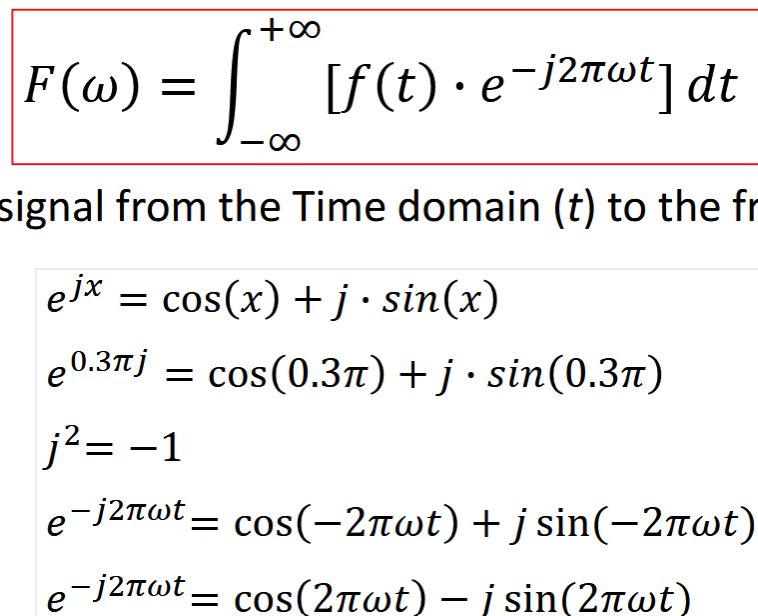
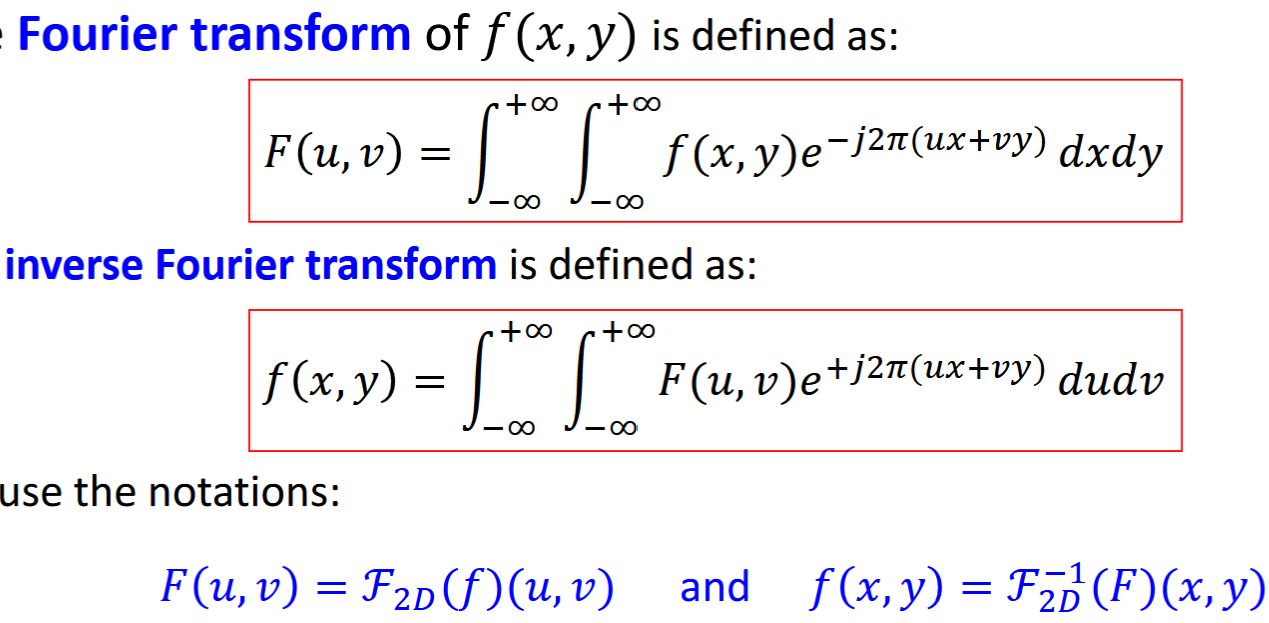
Fourier Transform
transforms signal from time domain to frequency domain
can be used to analyze spectral components, sep consider action of LSI at each sinusoidal frequency
F(u,v) = spectrum of f(x,y), sinusoidal composition at dif frequencies
Spatial vs. Frequency Domain
spatial - normal image, pixel coordinates and intensity values
frequency - FFT based frequency components, magnitudes and phases
Fourier Transform Spectrums
f(x,y) real valued, F(u,v) complex valued
FT of delta function = 1
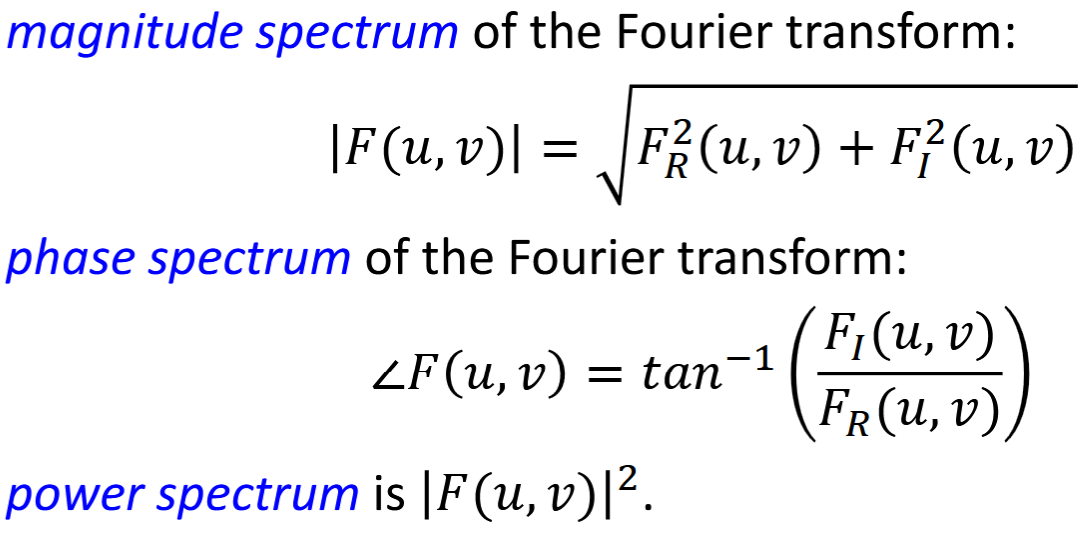
Fourier Transform Pairs
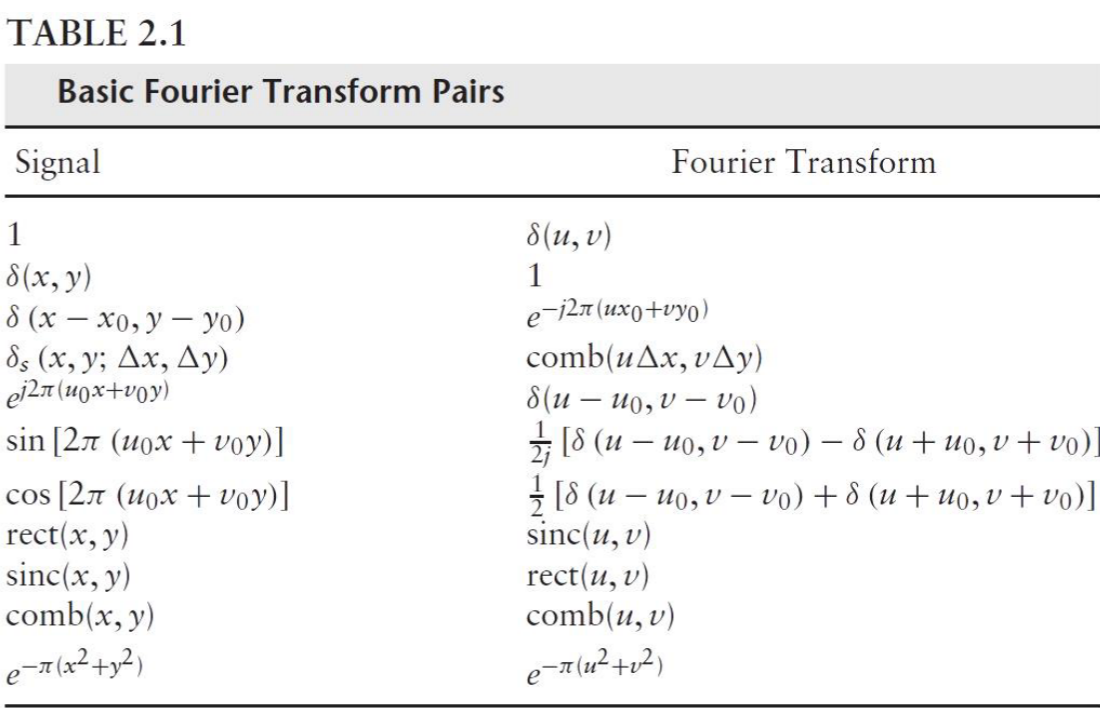
Fourier Transform Properties
Spacial Frequencies : Image Quality
LPF (low frequencies stay) - most of the image, blurred
HPF (high frequencies stay) - sharp edges of the image
slow signal variation = low frequencies
fast signal variation = sharper features, high frequencies
req high spatial frequency for fine structure high quality
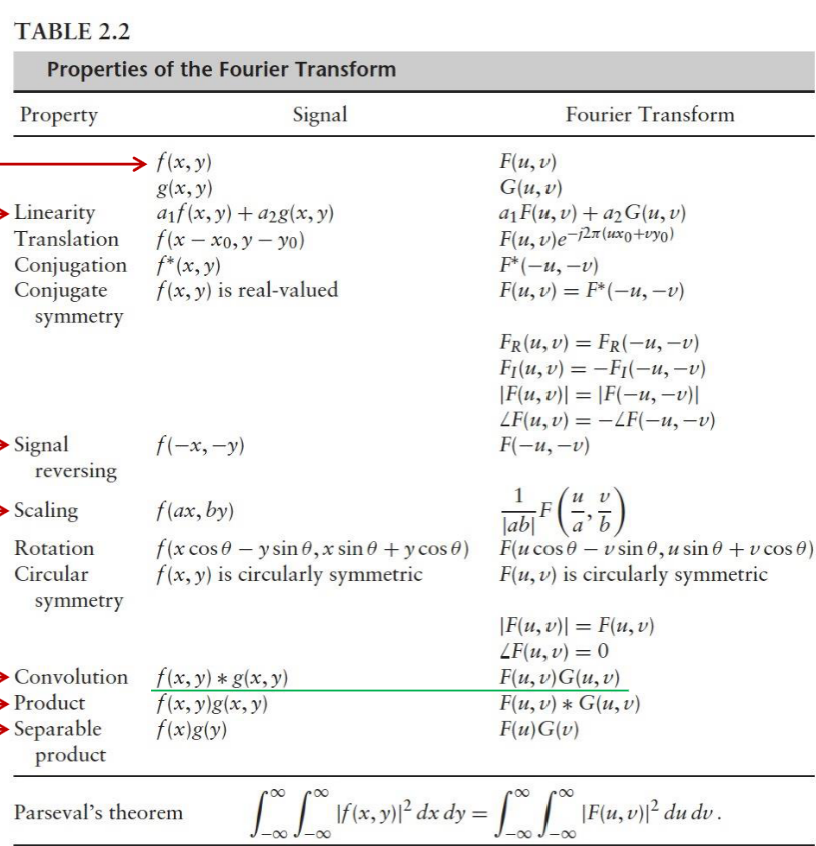
Convolution Theorem
convolution in spatial = multiplication in frequency
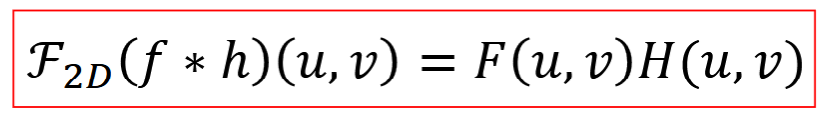
Parseval’s Theorem
total energy of a signal in the space domain = total E in frequency domain
FT & inverse are energy preserving
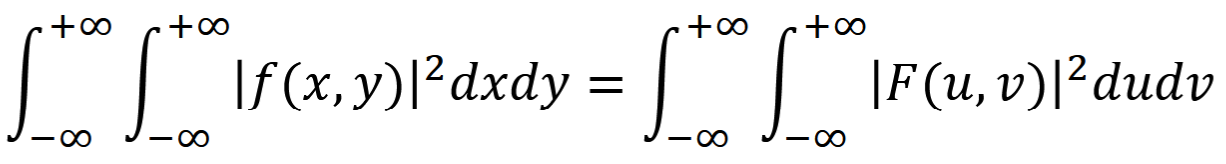
Transfer Function
Fourier transform of PSF, H(u,v)
optical transfer function
G(u,v)=H(u,v)F(u,v)
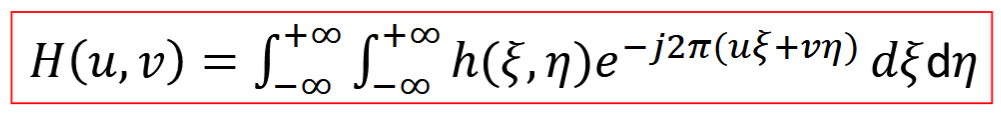
Cutoff Frequency
limited frequency bandwidth
filtering with it → signal smoothing (higher spatial frequencies eliminated via imaging, c1>c2)
Circularly Symmetric Signals
f_theta(x,y) = f(x,y) for every theta
Fourier transform is also circularly symmetric
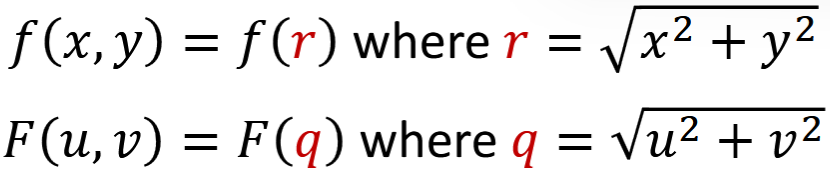
Hankel Transform
F(q)= H { f(r) }
Fourier transform of circularly symmetric 1 | 2D signals, including Gaussian
Image Quality Evaluation
contrast, resolution, noise (SNR), sampling (nyquist), artifacts, distortion, DA (sensitivity and specificity)
Image Quality Importance
image internal structures and functions of the patient body to diagnose abnormal conditions, guide therapeutic procedures, monitor the effectiveness of treatment
quality = degree to which an image allows accomplishment of these goals
Contrast
dif btwn image intensity of object and background, result of inherent object contrast within body
need high contrast for abnormality detection, preserve true object contrast
quantified with periodic signal with modulation, modulation (depth). mf<1, no contrast if mf=0
mg = modulation/contrast of the image = mf [H(u0,0)/H(0,0)]
![<ul><li><p>dif btwn image intensity of object and background, result of inherent object contrast within body </p></li><li><p>need high contrast for abnormality detection, preserve true object contrast </p></li><li><p>quantified with periodic signal with modulation, modulation (depth). mf<1, no contrast if mf=0</p></li><li><p>mg = modulation/contrast of the image = mf [H(u0,0)/H(0,0)]</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/bb13a953-45b7-4689-a72a-63b43a88885d.png)
Sinosoidal Object
f(x,y) = A + Bsin(2 pi u0 x)
fmax = A+B fmin=A-B mf=B/A
Modulation Transfer Function
MTF, mg/mf = |H(u,0)|/H(0,0)
quantifies degradation of contrast as function of spatial frequency. 0 <= MTF(u) <= MTF(0) = 1
better MTF = greater area under the curve
summed for subsystems by multiplication, overall is always less than single
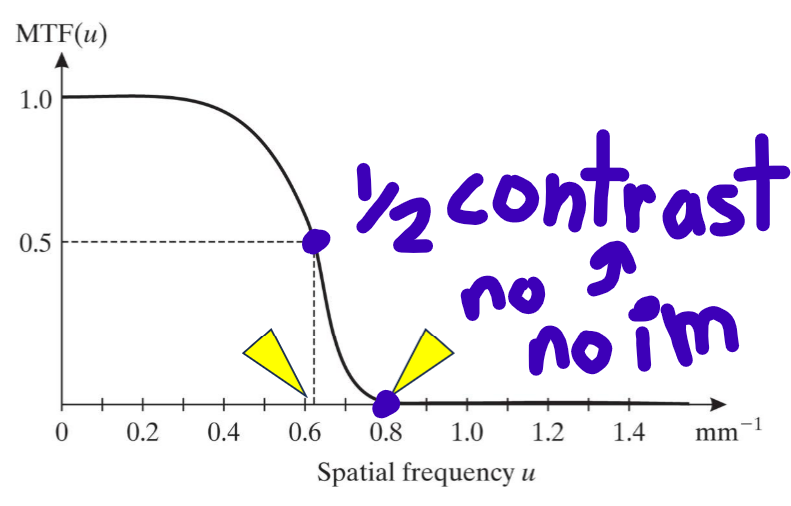
Local Contrast
difference btwn target and background
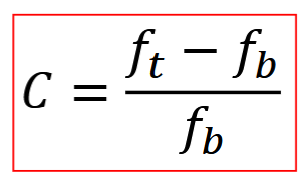
Contrast Mechanism
type and principle of contrast
CT - x-ray absorbtion coefficient of tissue
MRI - concentration of H atoms in tissue
Raman spectroscopy - chemical bond vibration
Fluorescence microscopy - fluorescence emission from fluorescent molecules
Resolution
capability to accurately depict two distinct events in space/time/frequency as separate corresponding to the spatial/temporal/spectral resolution
PET has high contrast and poor spatial resolution
can be quantified by the period of sinusoidal input. resolution = period of sine at 1/uc
Full Width at Half Maximum
full width of LSF/PSF profile at half the maximum value [mm], equals resolution
summed in square: R = sqrt(R1² + R2² +…+Rk²), heavily effected by large terms
![<p>full width of LSF/PSF profile at half the maximum value [mm], equals resolution</p><p>summed in square: R = sqrt(R1² + R2² +…+Rk²), heavily effected by large terms </p>](https://knowt-user-attachments.s3.amazonaws.com/5303c2f3-3997-4058-bd45-45db84450d23.png)
Bar Phantom
tool to measure resolution, image through the system to eval resolution
density of lines = line pairs per mm [lp/mm]
![<p>tool to measure resolution, image through the system to eval resolution</p><p>density of lines = line pairs per mm [lp/mm]</p>](https://knowt-user-attachments.s3.amazonaws.com/92880de3-95e6-4213-af19-adc8477b34ad.png)
Noise
any random fluctuation, unwanted char, numerical outcome of statistically random events
image quality decreases as noise increases over the signal
amplitude rep by SD, power rep by variance, a = average intensity, # of photons
CDF
cumulative distribution function
N (random variable) = number for random event, mathematically described by CDF
Pr(*) = probability; n = specific values of N.
PN(n) for continuous random variable = integral from - infinity to n of pN(u) du
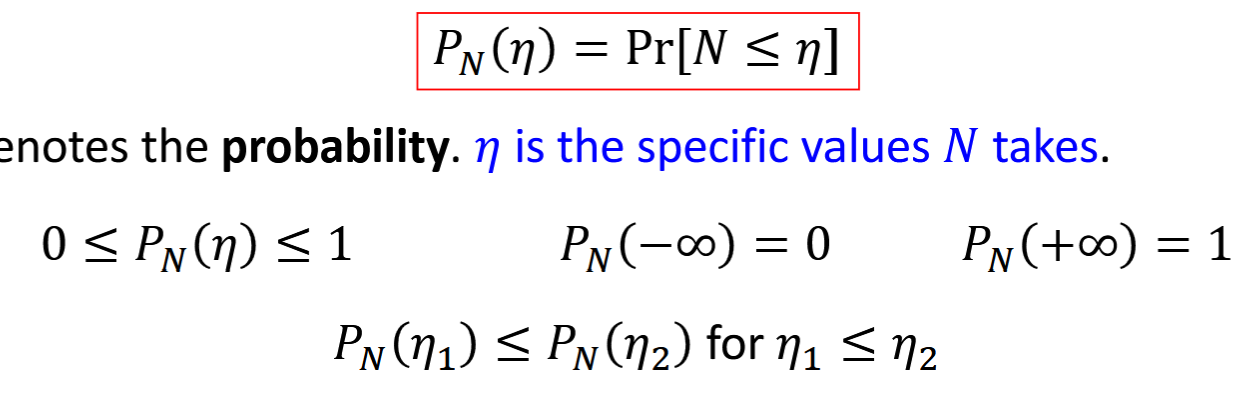
probability density function, gaussian and uniform
specifies continuous random variable. sums as convolution products
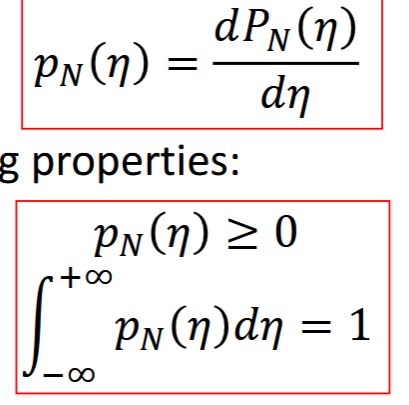
pmf
Probability mass function, discrete random variables (N discrete)
poisson
Stats for Noise
expected value = mean = uN E[N] = integral of n times pN(n) dn. sums
variance = oN² = Var[N] = E[(N-uN)²] = integral of (n-uN)² times pN(n) dn. sums
standard deviation = sqrt(variance)
integrals for continuous, summation for discrete
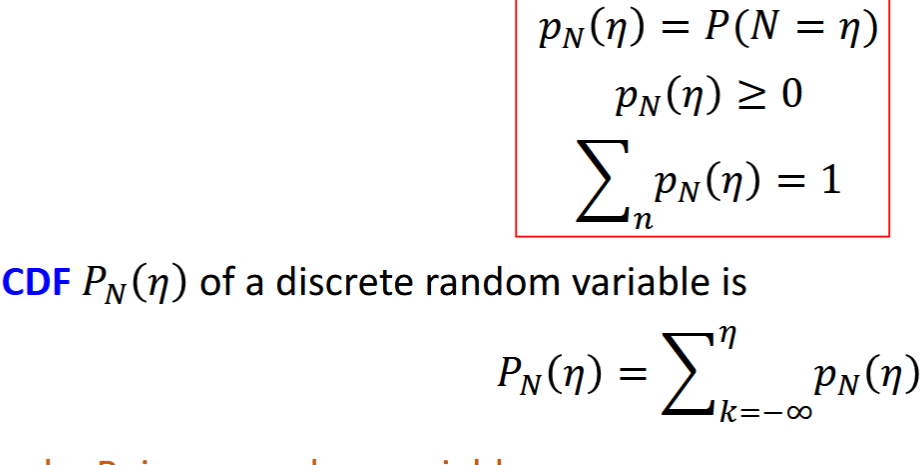
Uniform Random Variable
continuous, specific pdf
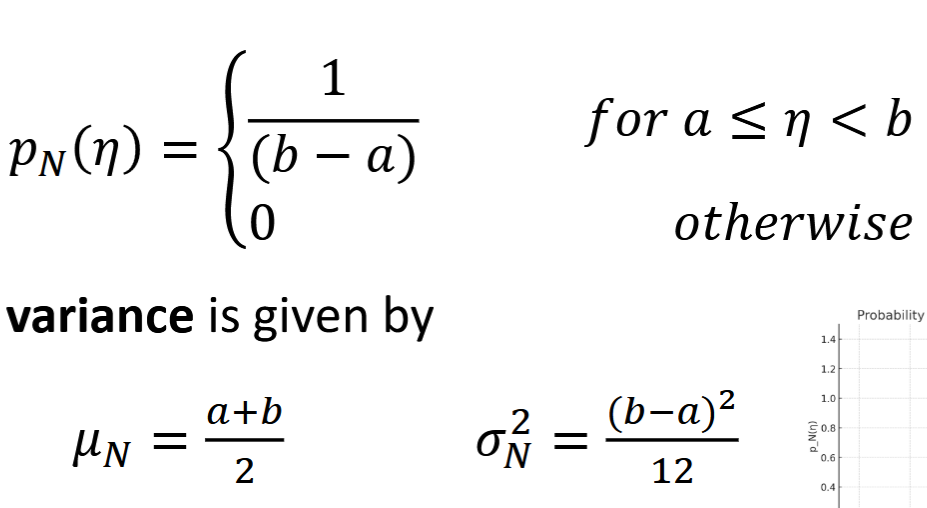
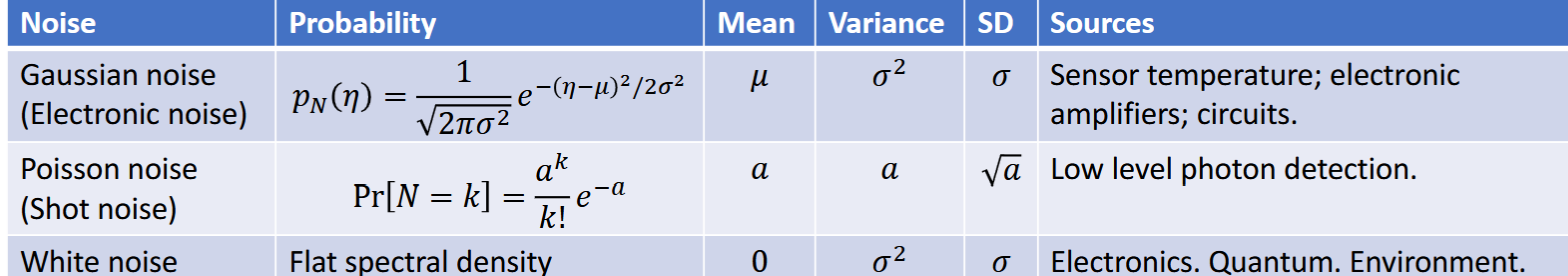
Gaussian Random Variable
continuous, natural to model noise with, based on pdf
Sensor temperature; electronic amplifiers; circuits
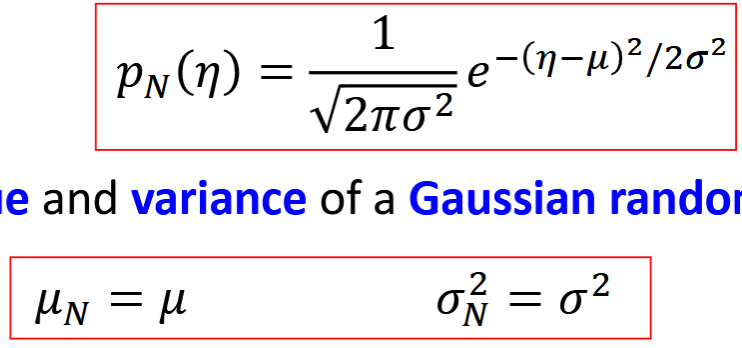
Poisson Random Variable
discrete, based on pmf, low level photon detection noise, a>0
graph has tail to the right. shot noise
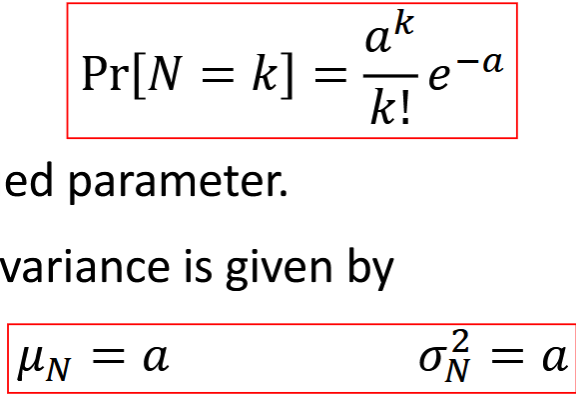
White Noise
equal intensity at different frequencies, constant power spectral density, Electronics. Quantum. Environment.
mean = zero, constant variance, serially uncorrelated Cov[N(t),N(s)]=0 for t not equal s
SNR
signal to noise ratio. prefer higher (output g more accurate to input f)
describes relative str of signal f with respect to noise N
blurring process reduces, higher noise reduces
Amplitude SNR
frequency amplitude / noise amplitude
Poisson = photons per unit area / standard deviation. SD = sqrt(mean) SNRa=sqrt(mean). more photons = higher SNR = higher im qual
SNR (in dB) = 20log(SNRa)
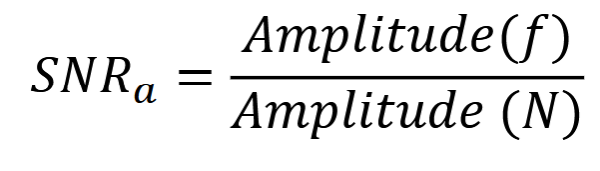
Power SNR
signal power / noise power
white noise: noise power = variance, mean = 0
SNR (in dB) = 10log(SNRp)
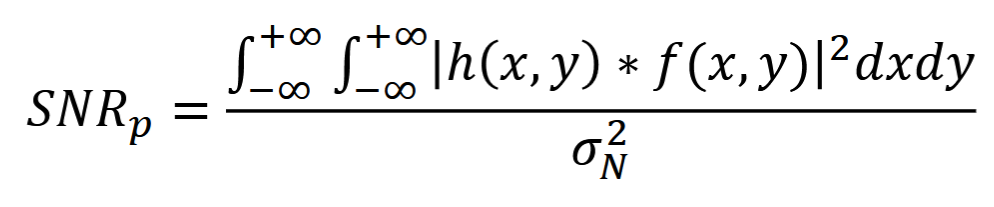
Frequency Dependent Power SNR
nonwhite noise, provides relation btwn contrast, resolution, noise, and image quality. up MTF = up im qual
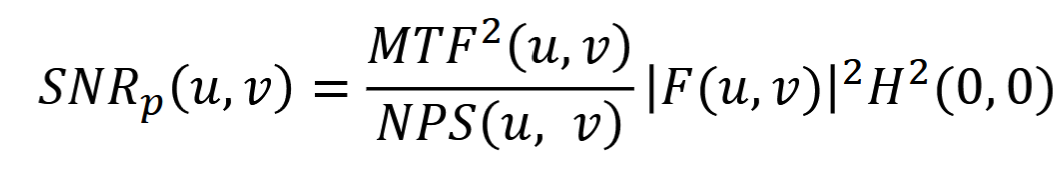
Differential SNR
A = area, ft = average image intensity within A, fb = average image intensity within A of background
SDn(A) = standard deviation of image intensity vals over area of the background
SNR (in dB) = 20log(SNRa)
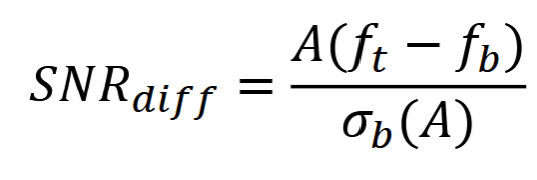
Sampling
∆𝑥 and ∆𝑦 are the sampling periods; 1/∆𝑥 and 1/∆𝑦 sampling frequency
𝑓𝑑 (𝑚, 𝑛) = 𝑓 (𝑚∆𝑥, 𝑛∆𝑦) ∆𝑥 <= 1/2U and ∆𝑦 <= 1/2V
Aliasing
artifact from low sampling, new high frequency patterns where none should exist
Nyquist Sampling Theorem- avoid aliasing by ∆𝑥(max) = 1/2U and ∆𝑦(max) = 1/2V. U=u0
Artifacts
features of an image that do not represent valid anatomical or functional information. obscure
a = motion, b = star from metal, c = detectors out of calibration, d = xray beam hardening
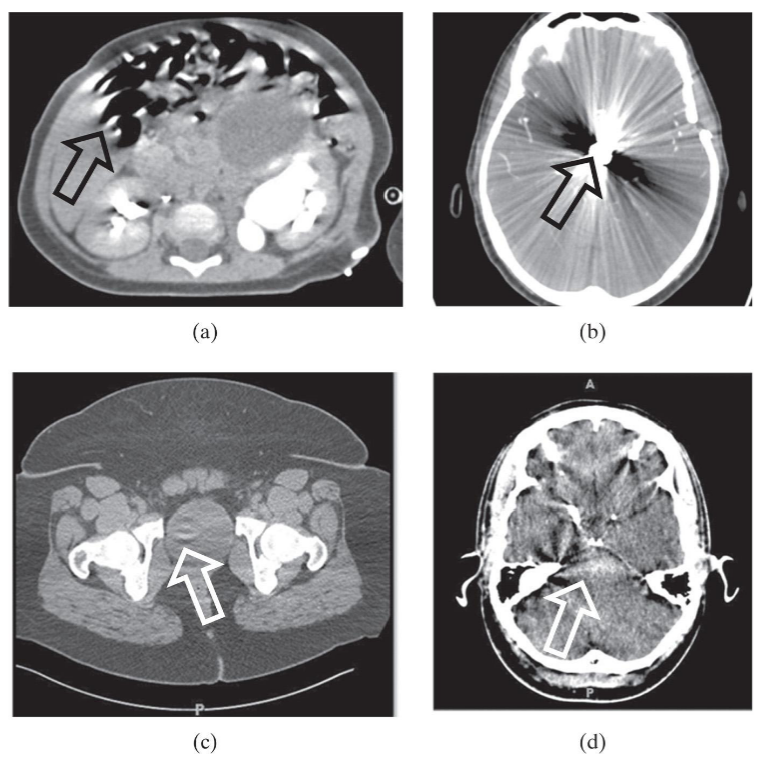
Distortion
inability of medical imaging system to give accurate impression of shape, size, or position of object
CT - dif size look same from dif distance from x-ray source. identical look different due to orientation
Accuracy
conformity to truth (free of error), clinical utility
in context of diagnosis, prognosis, treatment planning, treatment monitoring
quantitative accuracy and diagnostic accuracy
Quantitative Accuracy
numerical vals of given anatomic or functional image feature
error from bias (systematic reproducible difference, can be corrected) or imprecision (noise and random measure-measure variation)
Sensitivity
true positive fraction, fraction of patients with disease who the test calls abnormal
a/(a+c)
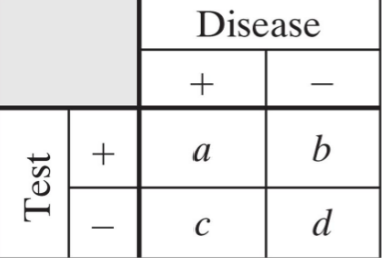
Specificity
true negative fraction, fraction of people without disease that the test calls normal
d/(b+d)
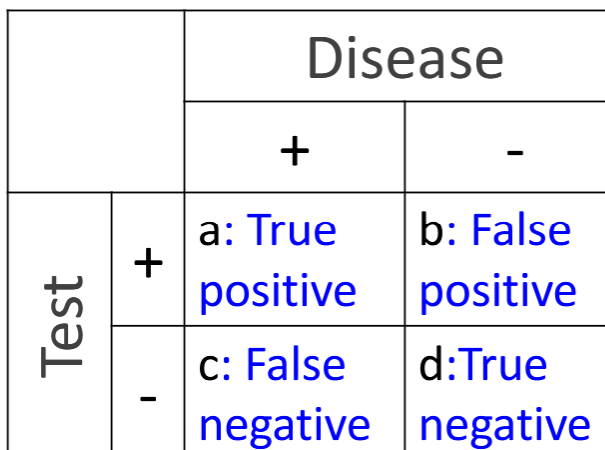
Diagnostic Accuracy
fraction of patients that are diagnosed correctly. max by max sensitivity and specificity
(a+d) / (a+b+c+d)
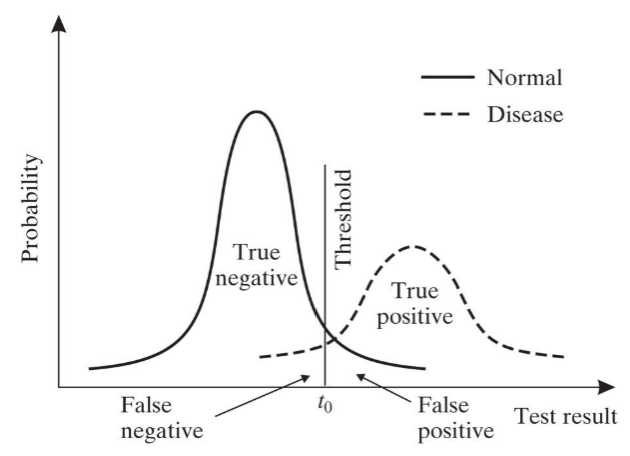
ROC Curve
lower threshold increases sensitivity and decreases specificity
plot of sensitivity (y) vs 1-specificity (x)
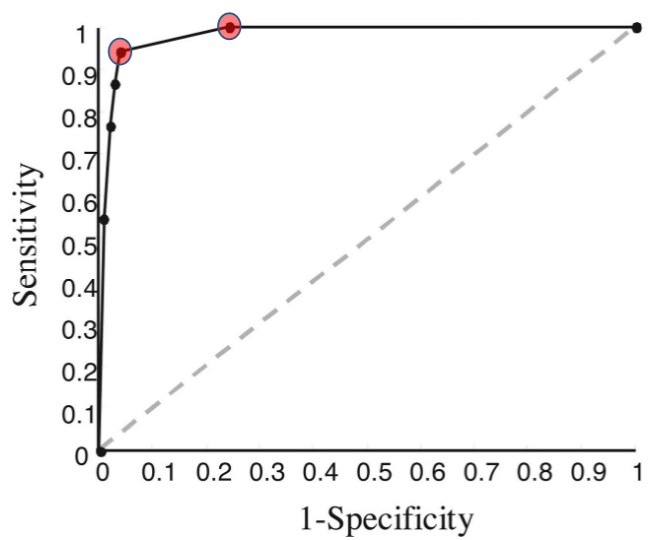
Prevalence
PR = (a+c) / (a+b+c+d) influences PPV and NPV
PPV = a / (a+b) called abnormal and have disease NPV = d / (d+c) called normal don’t have disease