Bio 2960-- Eukaryotic mRNA Processing
0.0(0)
Card Sorting
1/15
There's no tags or description
Looks like no tags are added yet.
Last updated 9:04 PM on 4/30/23
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
16 Terms
1
New cards
mRNA processing overview
mRNA is produced in the nucleus, and is also modified in the nucleus.
Main processing events:
1. 5’ mRNA capping
2. Cleavage and Polyadenylation
3. Intron Splicing
Main processing events:
1. 5’ mRNA capping
2. Cleavage and Polyadenylation
3. Intron Splicing
2
New cards
5’ mRNA capping
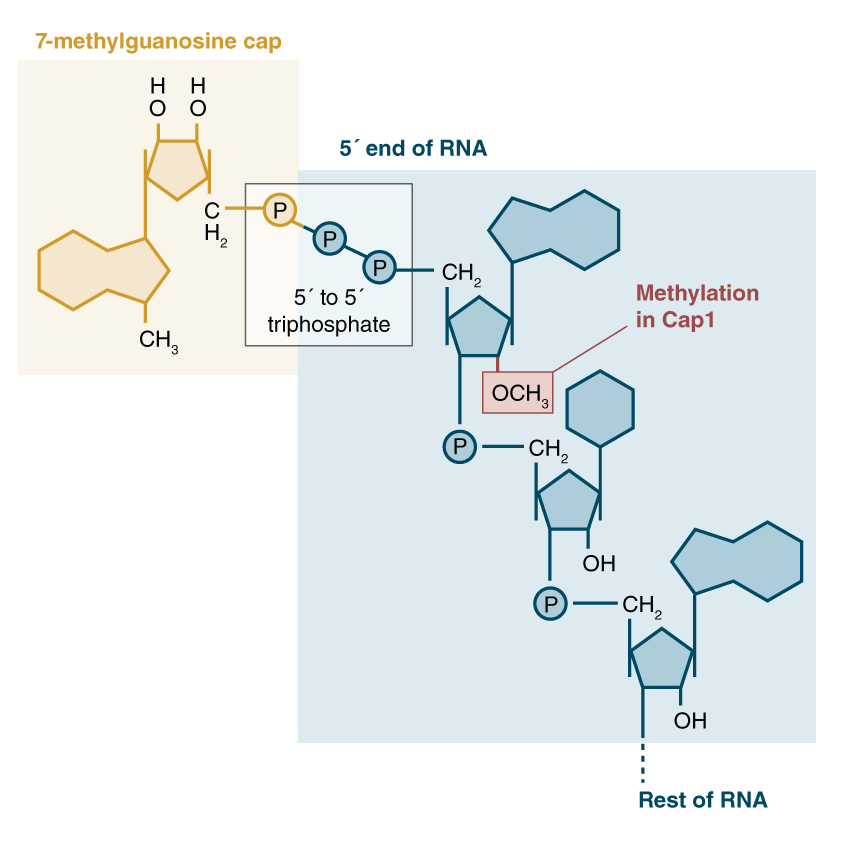
the cap is called m7G cap, and is a guanine nucleotide with a methyl group at the 7th residue. Caps binds to the 5’ end of mRNA.
1. Triphosphotase chops off a phosphate group from the 5’ end of the mRNA (recall the triphosphoate that hangs off every mRNA strand)
2. GTP is converted to **GMP**. This GMP is just the guanine nucleotide but with only one phosphate group. This Guanine is added to this 5’ end, and guanine and phosphate are connected via a **5’ to 5’ linkage.**
3. **Methyl group** added to 7th residue of guanine. Could also be added to ribose instead of guanine.
This cap protects RNA from exonuclease attacks and increases translational efficiency by allowing the ribosome to recognize the mRNA via that cap. It also binds to translation initiation factors.
1. Triphosphotase chops off a phosphate group from the 5’ end of the mRNA (recall the triphosphoate that hangs off every mRNA strand)
2. GTP is converted to **GMP**. This GMP is just the guanine nucleotide but with only one phosphate group. This Guanine is added to this 5’ end, and guanine and phosphate are connected via a **5’ to 5’ linkage.**
3. **Methyl group** added to 7th residue of guanine. Could also be added to ribose instead of guanine.
This cap protects RNA from exonuclease attacks and increases translational efficiency by allowing the ribosome to recognize the mRNA via that cap. It also binds to translation initiation factors.
3
New cards
RPII C terminal domain
these proteins responsible for creating the CAP hang onto RPII’s C terminal domain/tether, so they go with RPII as it transcribes. This allows the 5’ cap to be made as soon as the 5’ end is available. A stall factor in RPII gets RPII to stop for a little bit after the 5’ end is made so that the cap could be added.
4
New cards
Cleavage and Polyadenylation
1. Near the end of mRNA strand, there is an **AAUAAA** sequence that signals to an endonuclease that it needs to come make a cut a little downstream of that sequence. It then makes the cut.
2. At the region of that cut, **Poly A Polymerase** binds to RNA and adds 100-200 **A nucleotides** to mRNA. This creates a Poly A strand at the end of mRNA strands. Poly A Polymerase works **without a template.**
3. All Eukaryotes have 3’ AAA
Increases the stability of eukaryotic mRNA. Enhances translational efficiency.
5
New cards
introns vs exons
introns are the non-protein coding sequences that get cut out of mRNA. Exons are protein-coding that get joined together. Introns are usually larger than exons.
Introns make up of the majority of gene sequences.
Introns make up of the majority of gene sequences.
6
New cards
Intron Splicing locations
3 important locations in the intron:
1. At the **5’ splice site** (left end of the intron), there is a **GU** sequence
2. **Branch point**-- a point in the middle of the intron with an A nucleotide
3. At the **3’ splice site** (right end of intron), there is an **AG** sequence
This GU and AG define the beginning and end of the intron.
1. At the **5’ splice site** (left end of the intron), there is a **GU** sequence
2. **Branch point**-- a point in the middle of the intron with an A nucleotide
3. At the **3’ splice site** (right end of intron), there is an **AG** sequence
This GU and AG define the beginning and end of the intron.
7
New cards
Intron Splicing Process
Intron splicing done by **spliceosomes**-- Ribonucleo proteins that base pair with pre-mRNA and mark important regions for splicing. Made up of different subunits-- U1 and U2. These subunits are small nuclear RiboNucleo proteins.
1. U1 recognizes and bind to the GU at the 5’ splice site. Complementary base pairing follows.
2. U2 marks the branch point, and other subunits bind to the mRNA.
3. Once everything is bound, the actual cutting part of the spliceosome (U6) cuts the 5’ end of the intron, forming a loop/**lariat** because the 5’ end of intron is cut while 3’ end is still connected.
4. Then the 3’ end is cut, the lariat structure of introns is released, and the two exons are brought together.
1. U1 recognizes and bind to the GU at the 5’ splice site. Complementary base pairing follows.
2. U2 marks the branch point, and other subunits bind to the mRNA.
3. Once everything is bound, the actual cutting part of the spliceosome (U6) cuts the 5’ end of the intron, forming a loop/**lariat** because the 5’ end of intron is cut while 3’ end is still connected.
4. Then the 3’ end is cut, the lariat structure of introns is released, and the two exons are brought together.
8
New cards
transesterification reactions
Two transesterification reactions occur during intron splicing. One at the 5’ end of the intron, one at the 3’ end of the intron.
Recall that the 5’ splicing site is marked by GU, the 3’ is marked by AG, and the branch point is marked by A.
At the 5’ splice site, the **2’ OH group of A** attacks the phosphate group of the sugar-phosphate backbone at the **5’ splice site**. This cuts off the sugar phosphate backbone of the introns, and creates that **lariat**.
Now that that intron has been cut off, a **3’ OH of the first exon is free.** This OH attacks the 5**’ phosphate of the downstream exon**. This joins the two exons together and **releases the lariat**. The two exons become one continuous coding sequence.
Recall that the 5’ splicing site is marked by GU, the 3’ is marked by AG, and the branch point is marked by A.
At the 5’ splice site, the **2’ OH group of A** attacks the phosphate group of the sugar-phosphate backbone at the **5’ splice site**. This cuts off the sugar phosphate backbone of the introns, and creates that **lariat**.
Now that that intron has been cut off, a **3’ OH of the first exon is free.** This OH attacks the 5**’ phosphate of the downstream exon**. This joins the two exons together and **releases the lariat**. The two exons become one continuous coding sequence.
9
New cards
alternative splicing
there are different ways in which each mRNA can be cut, and this yields the formation of many different products from just one gene.
This can be done via varying 5’ and 3’ splice sites, mutually exclusive exons, and intron retention (not cutting out the exon at all)
This can be done via varying 5’ and 3’ splice sites, mutually exclusive exons, and intron retention (not cutting out the exon at all)
10
New cards
splicing regulation
**Repression**: RNA binding proteins bind to introns or exons at or near the consensus sequence and prevent subunits of the spliceosome from binding and thus cutting out introns
**Activation**: RNA binding proteins bind to splicing enhancer elements and promote spliceosome assembly and cutting out introns with poor 3’ or 5’ splicing sequence elements
**Activation**: RNA binding proteins bind to splicing enhancer elements and promote spliceosome assembly and cutting out introns with poor 3’ or 5’ splicing sequence elements
11
New cards
activator-dependent inclusion
If an exon has a weak 5’ splice site, it does not match the consensus sequence well, so it will require an activator to bind to it to be included in the final product. Activator binds to an exon slicing enhancer site on mRNA, and exon becomes well-defined and will be included in final product.
If activator is absent, exon remains poorly defined, and does not get included in final product. Would appear to the spliceosome as an intron.
If activator is absent, exon remains poorly defined, and does not get included in final product. Would appear to the spliceosome as an intron.
12
New cards
repressor-dependent exclusion
5’ and 3’ splice sites of exons are well-defined, but the repressor binds in a way that hides the 3’ splice site. This prevents the sNRNPs from binding to the 3’ splice site, and prevents that end of the exon from being included.
If this repressor is absent, sNRNP binds properly, exon remains well-defined, and is thus included in the mRNA.
If this repressor is absent, sNRNP binds properly, exon remains well-defined, and is thus included in the mRNA.
13
New cards
ribozyme
enzymes made of only RNA. There is a theory about the origin of life that hypothesizes that RNA was the only complex macromolecule around at the beginning of time since enzymes can be made completely of RNA.
Some mRNAs can cut out their own introns-- called self-splicing introns.
Some mRNAs can cut out their own introns-- called self-splicing introns.
14
New cards
translation in prokaryotes vs eukaryotes
same 3 steps and overall process-- Initiation, Termination, Elongation. Still uses ribosomes, tRNAS, amino acids, etc.
However, eukaryotic ribosomes have larger rRNA, an extra subunit (18s subunit), and more proteins. Overall they are just more complex.
They also differ in the specific steps of initiation.
However, eukaryotic ribosomes have larger rRNA, an extra subunit (18s subunit), and more proteins. Overall they are just more complex.
They also differ in the specific steps of initiation.
15
New cards
Initiation of Translation in eukaryotes
1. Eukaryotic Initiation Factor (EIF) recognizes the 5’ cap and binds to 5’ end of mRNA.
2. Initiation factors and small subunit of ribosome come together at start codon
3. 40s subunit of ribosome brings in the first tRNA. It scans the mRNA for the first AUG. The first AUG it encounters becomes the start codon.
4. Once start codon is found, initiation factors leave, other ribosomal subunit joins, and translation begins
16
New cards
translational regulation
Translation in eukaryotes can be regulated by microRNAs.
1. These miRNAs are transcribed by RPII from noncoding regions of DNA.
2. Initially, they are **pri-microRNA**s in a hairpin loop structure.
3. Then, **Drosha** comes and releases this hairpin structure. This becomes **pre-microRNA**.
4. This pre-microRNA exits the nucleus and goes to the cytoplasm, where dicer cuts off the loop and releases the **microRNA** into the cytoplasm.
microRNA then binds to RNA silencing proteins and forms an **RNA-Induced Silencing Complex**. This complex includes one strand of miRNA, and this strand forms complementary base pairing with the mRNA strand. This prevents that mRNA strand from being translated because the base pairing is imperfect.
1. These miRNAs are transcribed by RPII from noncoding regions of DNA.
2. Initially, they are **pri-microRNA**s in a hairpin loop structure.
3. Then, **Drosha** comes and releases this hairpin structure. This becomes **pre-microRNA**.
4. This pre-microRNA exits the nucleus and goes to the cytoplasm, where dicer cuts off the loop and releases the **microRNA** into the cytoplasm.
microRNA then binds to RNA silencing proteins and forms an **RNA-Induced Silencing Complex**. This complex includes one strand of miRNA, and this strand forms complementary base pairing with the mRNA strand. This prevents that mRNA strand from being translated because the base pairing is imperfect.