37 - DNA Translation
1/45
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
46 Terms
What is the role of aminoacyl-tRNA synthetase?
Catalyzes the covalent attachment of amino acids to the 3' end of tRNA molecules.
Aminoacyl-tRNAs are high-energy molecules, and the activation of an amino acid by this enzyme requires ATP.

How does aminoacyl-tRNA synthetase bind the correct amino acid?
It binds ATP and the correct amino acid, and then binds the specific tRNA based on structural features (such as the anticodon or acceptor stem). Some of them can proofread.
Describe the steps of translation.
Aminoacyl-tRNA binds to the A site, requiring GTP, eEF-1.
Peptide bonds form in the A site via peptidyl transferase in the large subunit of the ribosome.
Translocation of the ribosome moves it 3 nucleotides along the mRNA, requiring GTP, eEF-2.
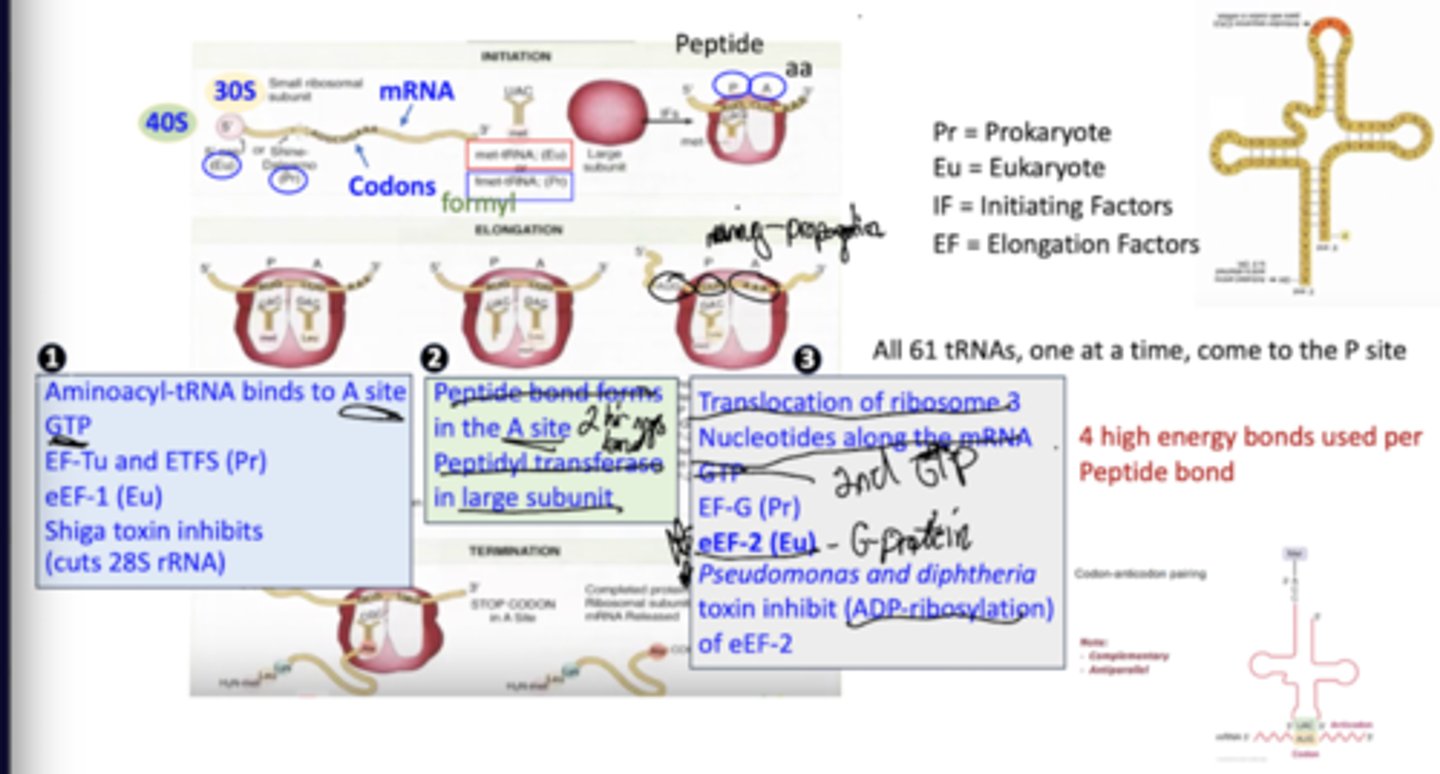
Where do tRNAs come to the ribosome? In what order?
All 61 tRNA come to the A site of the ribosome one at a time, in the order dictated by the mRNA.w
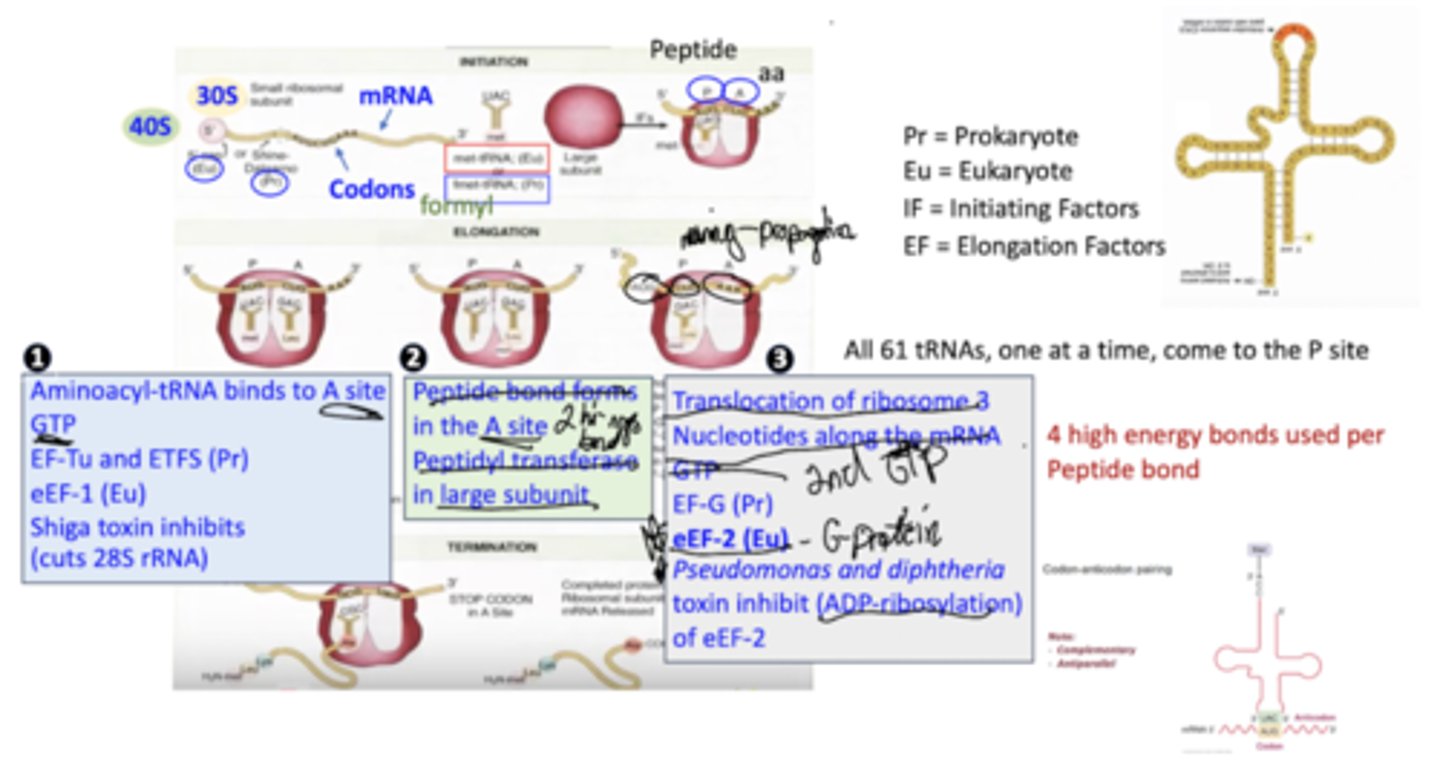
How much energy is used to form peptide bonds?
4 high energy bonds are used to form each peptide bond
What are shiga toxins?
They inhibit the binding of aminoacyl-tRNA to the A site by cutting 28S rRNA.
What is the effect of pseudomonas and diphtheria on translation?
They inhibit of eEF-2 by ADP-ribosylation and, therefore, inactivation.
What is the effect of the ahminoglycosides, streptomycin and gentamycin, on translation?
They inhibit the interaction of the 30S prokaryotic ribosomal subunit with the Shine-Dalgarno sequence.
What is the effect of linezolid on transcription?
Inhibits initiation of transcription by binding to the large subunit (50S) and prevent it from forming the ribosome.
This drug is a last resort antibiotic for patients resistant to all other antibiotic treatments.
What is the effect of tetracycline on transcription?
It inhibits the binding of aminoacyl-tRNA to the A site
What is the effect of chloramphenicol on transcription?
It inhibits peptidyl transferase, preventing the formation of peptide bonds. This leads to grey-baby syndrome, which doesn't sound real but it probably is.
What is the effect of macrolides on transcription?
Binds to the 50S subunit, inhibiting translocation
What is the effect of clinamycin on transcription?
It is NOT a macrolide, but it inhibits the 50S translocation as well.
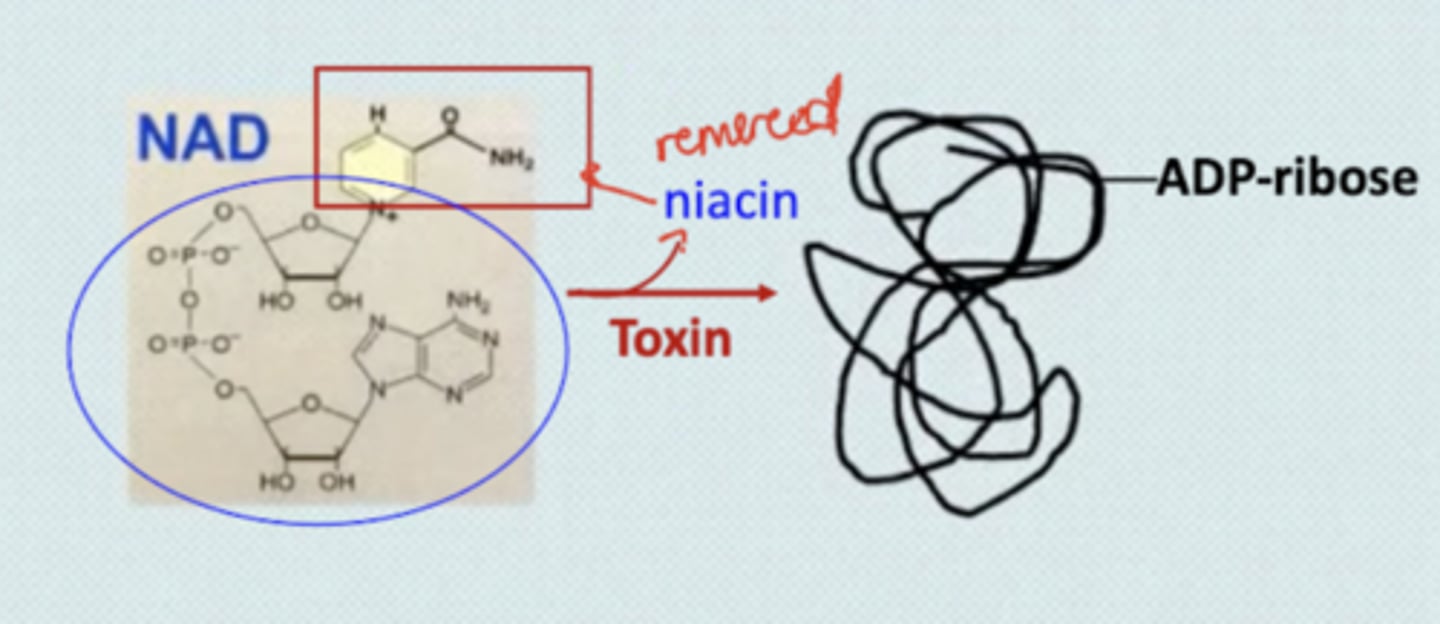
How do toxins, such as pseudomonas and diphtheria affect NAD?
They cause the loss of niacin by replacing it with an ADP-ribose
What is the fate of old proteins?
Old proteins are turned over, depending on a variable half life (enzymes have shorter half lives). They are tagged with ubiquitin (req ATP) and brought to the proteasome to be completely destroyed. Ubiquitin is recylced by E3 ubiquitin ligase.
What types of proteins assist in protein folding?
Chaperone proteins or heat shock proteins.
How are heat shock proteins related to cancer?
Overlord Kashfi research here. FoxM1 is a master regulator of the cell cycle and misfolding can lead to cancer.
What is the role of the N-terminal hydrophobic signal sequence?
It is a 10-15 hydrophobic amino acid sequence that prevents misfolding. It involves signal recognition and assists in peptide threading through the ER lumen.
What is the difference between O- and N- glycosylation?
N- glycosylation is the addition of sugar to the nitrogen of asparagine. This is co-translational and happens in the ER. O- glycosylation is the addition of sugars to the -OH of serine or threonine. This is post-translational, and happens in the golgi.
What post-translational modification signals a protein to the lysosome?
Phosphorylation of mannose by phosphotransferase (PO4 to mannose).
What proteins are NOT glycosylated?
Albumin, insulin, and glucagon.
What is I-Cell Disease?
Inclusive body disease. This is a result of defects in phosphotransferase. (tags for lysosomal proteins) Enzymes are thus secreted, leading to generalized inflammation.
Inclusion bodies are often found in fibroblasts, mannose-6-phosphate inclusions will be found, and coarse facial features will be present, as well as joint pain and psychomotor retardation in a young child.
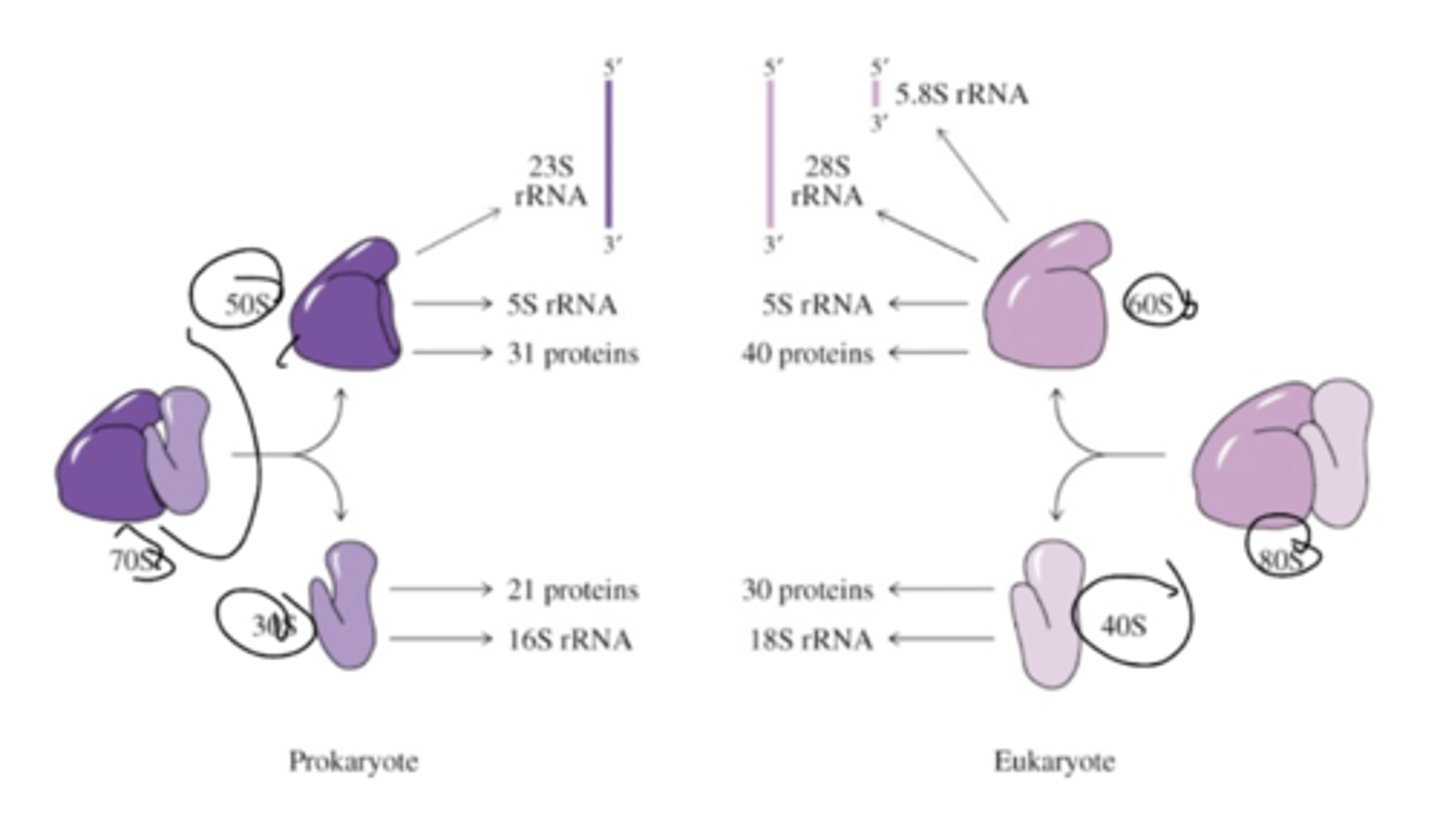
What are some characteristics of ribosomes?
The ribosome complex (30S and 50S to make 70S, or 60S and 40S to make 80S) moves 5' to 3' along the mRNA template (N --> C synthesis of the polypeptide)
What are some characteristics of the initiation complex?
Assembles at the first mRNA codon and disassembles at the termination step.
What are the components of the translation complex?
Ribosomal subunits, mRNA template, initiator tRNA, and protein initiating factors
What is initiatior tRNA?
The tRNA corresponding to AUG (methionine). Each cell contains two methionyl-tRNAMet that recognize it (second only for internal AUG segments).
Bacteria have N-formylmethionyl-tRNAfMet (has a formyl (COOR) group). Eukaryotes have methionyl-tRNAiMet.
What are initiation factors?
Required to form the initiation complex. IF-1, 2, and 3 in prokaryotes, and eIF in eukaryotes.
What is the significance of eIF-4?
Eukaryotic IF-4, aka the cap binding protein (CBP), that binds to the 5' end (7-methylguanylate cap) of the mRNA.
What is the role of the preinitiation complex?
It consists of the 40S ribosome, aminoacylated initiator tRNA, and other factors. It searchers for the initiator codon, and once found, the 60S subunit binds to complex the complex.
Where do the initiation complexes assemble?
In prokaryotes, 30S ribosome binds to region of mRNA (Shine-Dalgarno) upstream of initiation sequence. The S-D also binds complementary base sequence at the 3' end of the 16S rRNA (double-stranded RNA structure binds to mRNA to ribosome.
Describe the process of chain elongation.
Initiator tRNA begins in the P site, with the A site ready to receive aminoacyl-tRNA. There are three steps: positioning correct aa-tRNA in site A, formation of peptide bond, and shifting mRNA by one codon.
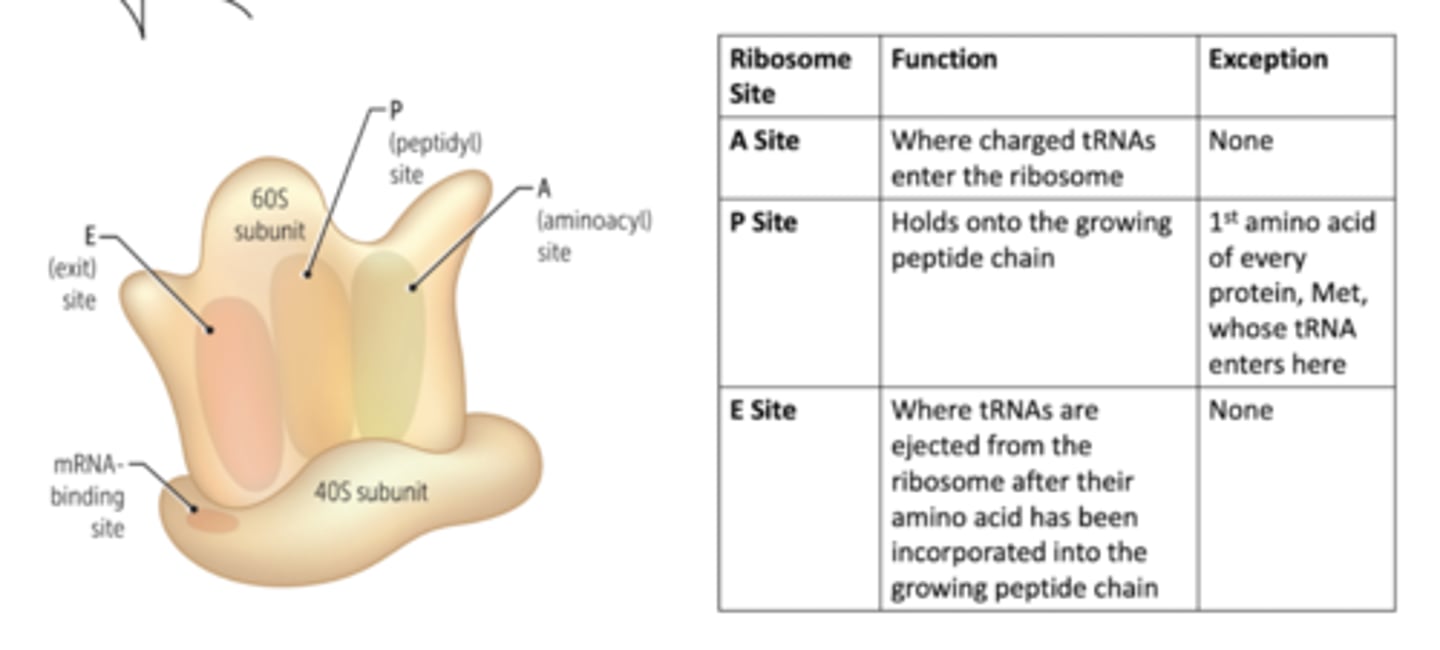
Where does peptidyl transferase activity take place?
Peptidyl transferase activity contained within LARGE RIBOSOMAL SUBUNIT (substrate binding site in 23S rRNA and 50S ribosomal proteins with catalytic activity from 23S rRNA.
What happens during translocation during chain elongation?
new peptidyl-tRNA moved from A to P site, and mRNA shifts by ONE CODON. Deaminoacylated tRNA shifted from the P site to the E site (exit).
What is EF-2?
EF-2 (eukaryotic elongation factor) - G protein requiring GTP and a target for toxins
What energy is required for protein synthesis?
Four phosphoanhydride bonds are cleaved for each amino acid added to a polypeptide chain
Activation: Two Pi bonds (ATP → AMP + 2Pi)
Chain Elongation: Two PI bonds (2 GTP → 2 GDP + 2Pi)
What is the role of puromycin in protein synthesis?
It resembles 3' end of aminoacyl-tRNA and can enter A site of ribosome.
Peptidyl-puromycin formed bound weakly to A site and dissociates, terminating protein synthesis -- PROKARYOTES
How is the synthesis of ribosomal proteins regulated?
It is regulated at translational level
Ribosomal protein genes encode one ribosomal protein that inhibits translation of its own polycistronic mRNA by binding near initiation codon of mRNA
What are some posttranslational modifications?
Occur either before polypeptide chain is complete or after
Deformylation of N-terminal residue (prokary)
Removal of N-terminal methionine residue
Formation of disulfide bonds
Cleavage by proteinases
Phosphorylation or acetylation
Describe the key sites of ribosome function?
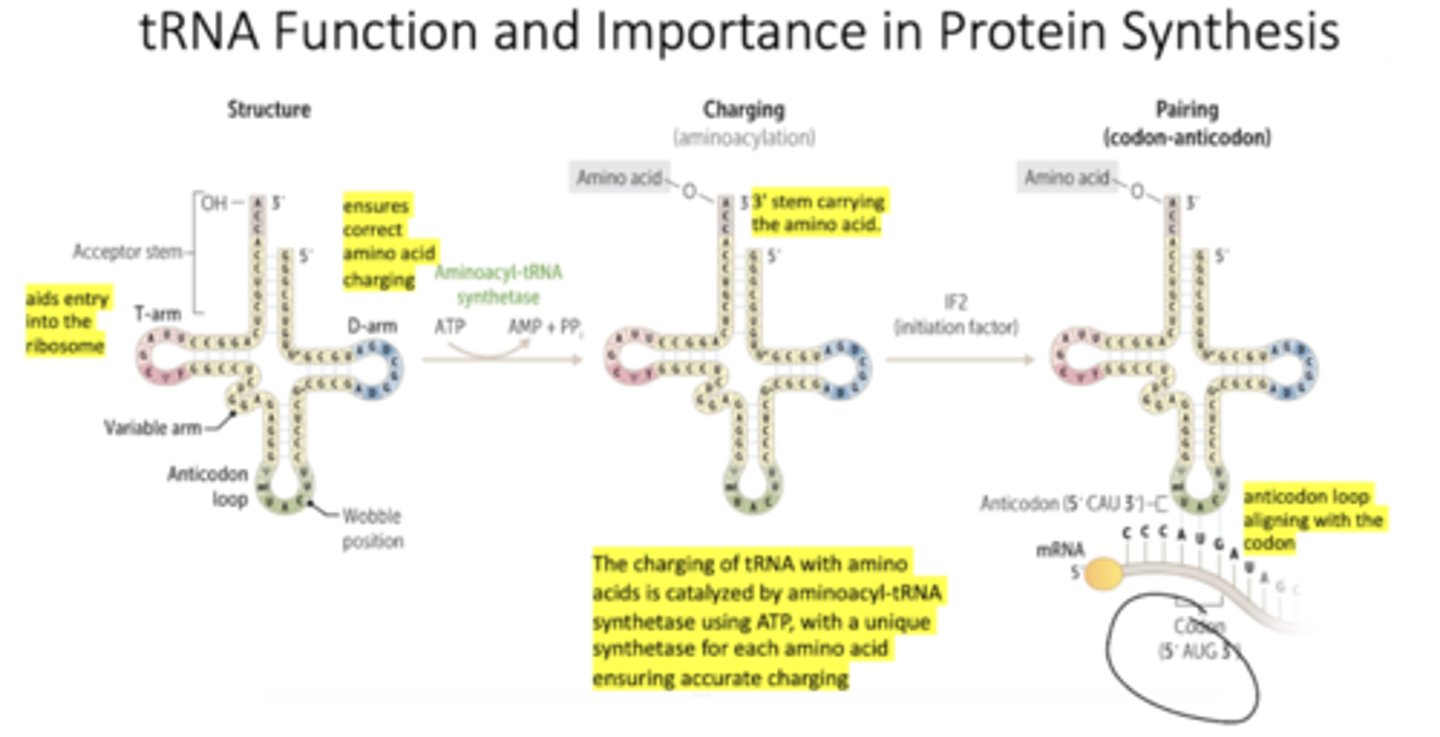
What are some characteristics of tRNA function?
What is the format of the genetic code?
The genetic code consists of three letter codons that don't overlap, with tRNA adapters that translate between mRNA and amino acids.
The reading frame is each potential starting point for interpreting the 3 letter code
This reading frame consists of 64 possible codons, 61 of which actually encode for protein (the open reading frame not containing stop codons), and 3 of which are stop codon (U Go Away, U Are Away, U Are Gone; UGA, UAA, UAG)
The start codon is AUG or ATG in DNA
What are some characteristics of the genetic code?
Characteristics of the genetic code:
Unambiguous, with each codon corresponding to one amino acid
Multiple codons for most amino acids (degenerate; synonymous codons specify same amino acid)
What are some basic characteristics of tRNA?
tRNA interpret genetic code; at least 20 tRNA per cell
Cloverleaf shape
Named for the amino acid they carry (tRNAPhe)
5' anticodon position has flexibility in base pairing (wobble position)
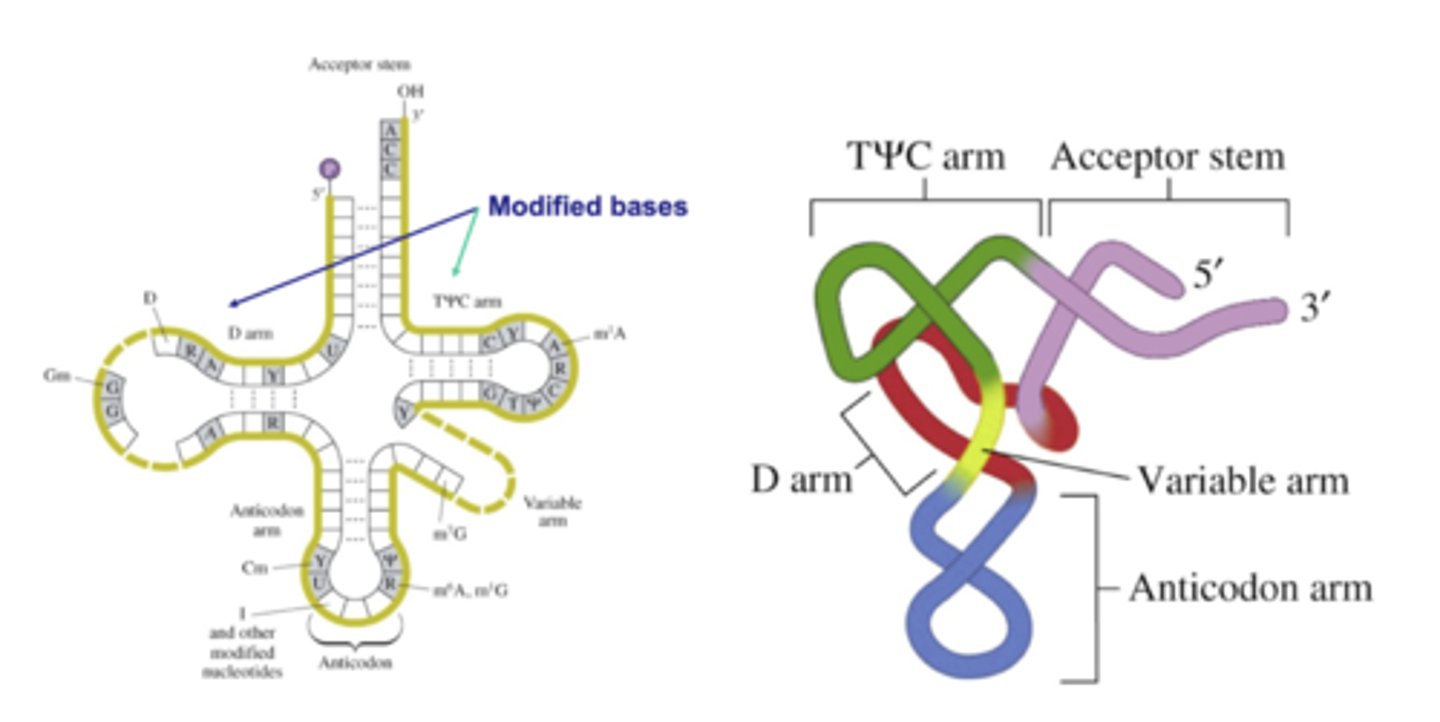
What are the different arms of tRNA responsible for?
Acceptor stem: amino acid becomes covalently attached to tRNA at the 3' end of the stem
Anticodon arm: contains anticodon that binds to complementary codon in mRNA
T epsilon C arm: contains thymidylate (T) and pseudouridylate (epsilon) followed by C
D arm: contains dihydrouridylate (D)
Variable arm: ranges from 3-21 nucleotides
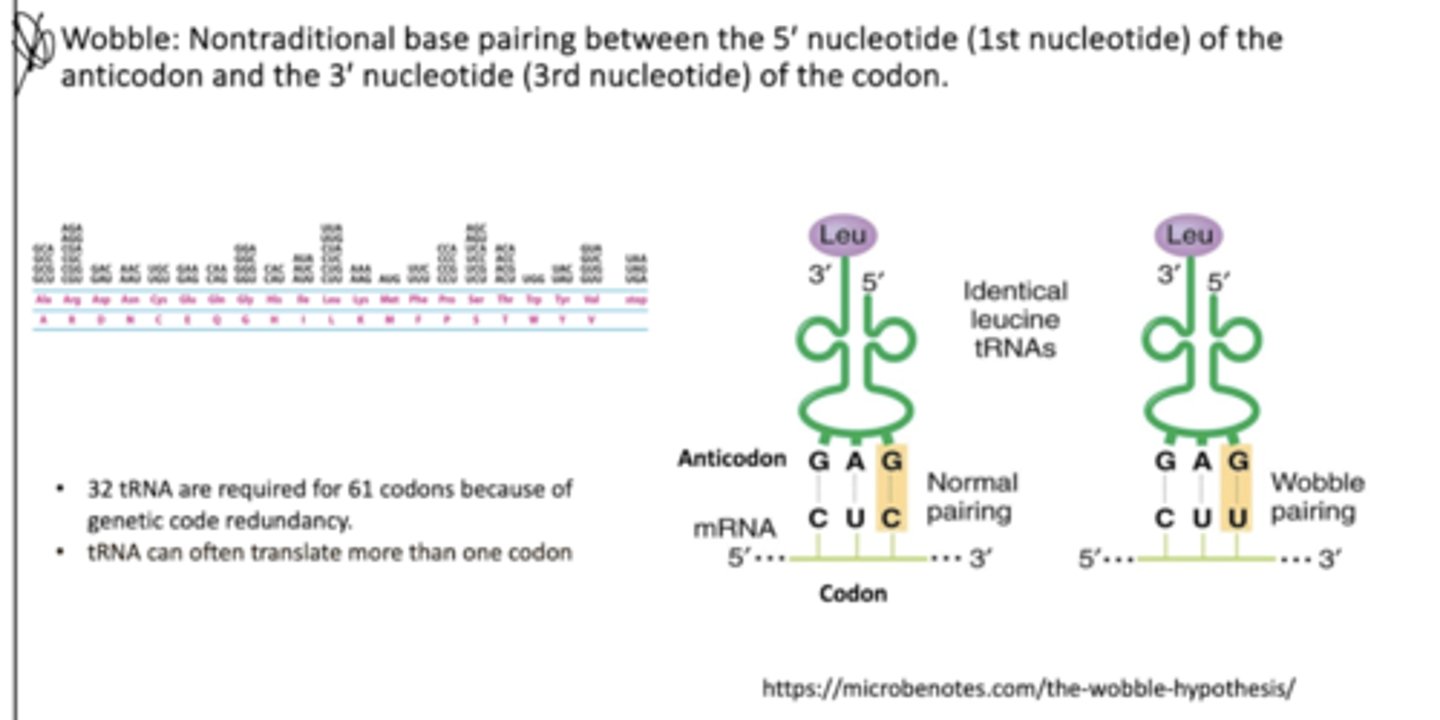
How is the codon recognized by the anticodon?
Wobble allows tRNA to recognize more thna one codon
Isoacceptor tRNA molecules: different tRNA molecules bind the same AAs
Defined by roman numerals or codons (tRNAIAla, tRNAIIAla, or tRNAGCGAla)
What is the wobble hypothesis?