Lecture 7 - Eukaryotic Transcription Termination, Export, and Methods of Analysis
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
25 Terms
Nuclear Pore Complex
large, multiprotein complexes embedded in the nuclear envelope allow the movement of the TFs between nucleus and cytoplasm
small neutral proteins can passively pass through the pore
very large proteins and viruses can be translocated across the nuclear envelope by vesicular trafficking
What can be escorted through the nuclear pore?
charged proteins, RNAs, and some large proteins
proteins can be targeted to the nucleus by a nuclear localization signal (NLS)
proteins can be repeated moved in and out of the nucleus by, and the exit would require a nuclear export signal (NES)
Karyopherins
regulate import and export
NES and NLS defined a consensus aa sequence that are recognized by proteins, exportins and importins, respectively (NLS is retained in the protein) → typically lysine and arginine rich
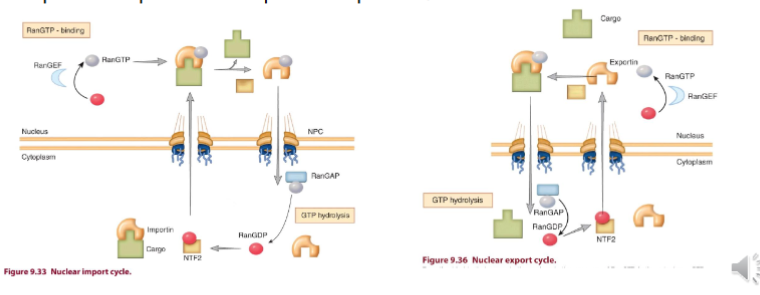
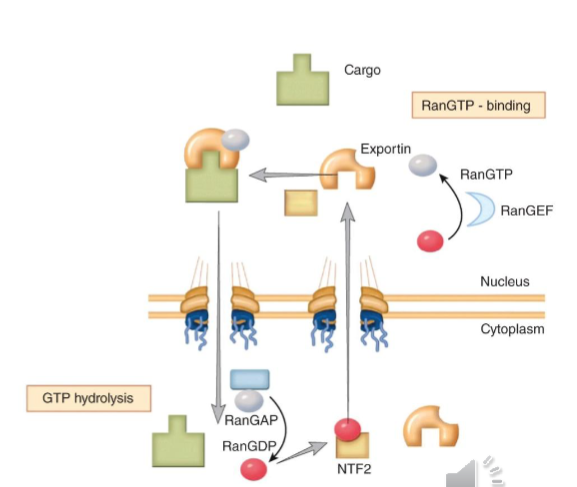
Simplified Classical Model of Karyopherins
complex of importin-a and importin b-1 bind to NLS of cargo protein
complex of importin-a and importin b-1 with the cargo docks at the nuclear pore complex on the cytoplasmic side
cargo is exported across NPC
once on nuclear side of NPC, cargo is disengaged from the importin complex facilitated by Ran
RanGEF exchanges the nucleotide in Ran-GDP with GTP making Ran-GTP → binds to importin complex causing an allosteric change that results in the release of the cargo
Ran-GTP-Importin complex moves out of the nucleus, with help from exportin
once outside the nucleus, RanRAP hydrolyzes RANGTP making RanGDP and process repeats once importins are recycled
RanGTP
concentrations are high in the nucleus
is essential for assembly of the Export Complex
is essential for disassembly of the Importin Complex
Termination in Brief
transcription: initiation, elongation, proof-reading, termination
in E. coli there are Rho dependent and independent mechanisms of termination; premature termination is transcriptional attenuation
eukaryotic termination is less understood but it is necessary to recycle RNAP II and prevent interference between adjacent genes
RNAP I requires a polymerase-specific termination factor that binds to a specific DNA sequence
RNAP II terminates following 3’ processing of the transcript - polyadenylation by protein complex that is carried by the phosphorylated CTD tail and binds to elements in 3’ UTR of mRNA
RNAP III terminates after transcribing a series of U residues but does not require a stem-loop structure
DNA
gene: entire DNA sequence necessary for the synthesis of RNA molecule or functional polypeptide which includes coding and regulatory regions
information carried by a gene is converted into observable product (the definition of gene expression)
Assessing Gene of Interest
can be done by observing abundance of RNA transcribed from the gene OR observing the amount of protein translated from this mRNA
questions to ask:
is the gene of interest transcribed: yes no
If yes – is it transcribed more then control: yes no
If yes – did we find more of the accumulated transcript (mRNA):
yes no (as mRNA is getting degraded...)If yes – is transcript viable and carried to the ribosomes for translation:
yes no (as mRNA continues to get degraded...)If yes – is the protein produced: yes no (unfavourable conditions?)
If yes – is the protein properly folded: yes no
If yes – is protein properly distributed and/or functiona
The method used depends on whether you are trying to detect ___, ___. or ____.
DNA; RNA; protein
nucleic acid hybridization is a means for detecting complementary in DNA or RNA samples
Southern blotting detects DNA (capital S)
northern blotting detects RNA
protein-protein interaction is a means for detecting proteins with other proteins (antibodies)
western blotting detects protein
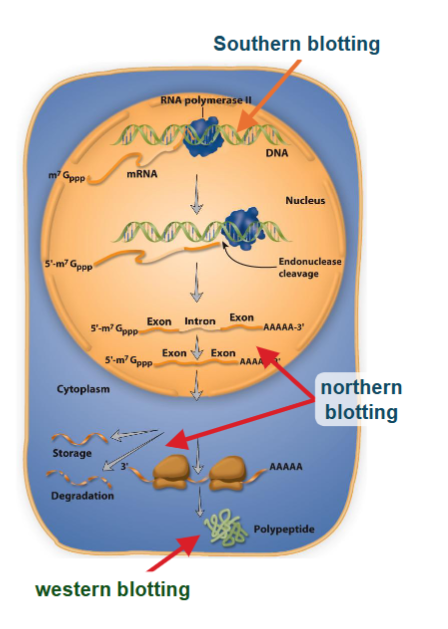
Detecting Specific Gene or Product
DNA/RNA needs a labeled hybridization probe - 10mer-100mer of nucleotides complementary to gene of interest (DNA; Southern, RNA; northern)
protein needs a labeled antibody
generally speaking: target is unknown sequence or protein; probe is known sequence or antibody
What does it mean to be labeled?
has to be something identifiable, detectable, measurable
hybridization probe or antibody:
use radioactive atoms in nucleotides (usually T for DNA and U for RNA probes)
use covalently attached fluorescent molecules, variety of enzyme conjugates
incorporated into probe during synthesis
method for detection (measuring) radioactivity, or light, or enzymatic product
labeled probe will hybridize with target and be detected
Source of Labeled Probe
DNA
whole or partial coding sequence for the gene of interest from same organism OR a different organism OR sequence from the same family as gene of interest
generally use labeled nucleotides to do de novo RNA or DNA synthesis from one of the above DNA sources as a template
homologous probe: probe and target sequences are perfect match (100% complementary)
heterologous probe: when probe and target are not 100% complementary, some degree of mismatching of bases
protein:
target protein in enough quantity for antibody production (monoclonal, polyclonal)
labelling with enzymes or fluorescent tags can be added after antibody production and purification
Level of Complementarity of Probe with Target
depends on the template that was used to make the probe
% of sequence identity (homology) between target sequence and prob determined the hybridization conditions
higher homology = higher stringency
hybridization conditions (salt, temperature) change the min homology needed for hybridization to the successful between target and probe
stringency considerations are also affected by probe size and actual sequence used as a probe or targeted
ultimate goal is for nucleic acid hybridization is to bind the probe to the target in a controlled manner
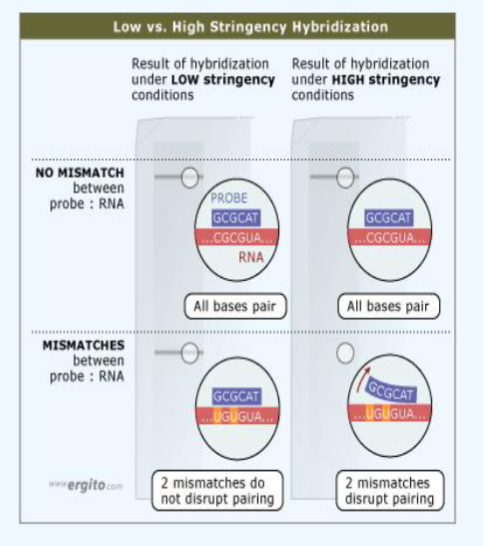
Stringency
controlling conditions under which hybridization is done and minimum homology needed for it to be successful → a measure of the tolerance for mismatches between target and probe
% identity between target and probe determines stringency
higher similarity = higher stringency, less change for mismatch
lower similarity = lower stringency, more chance for mistake
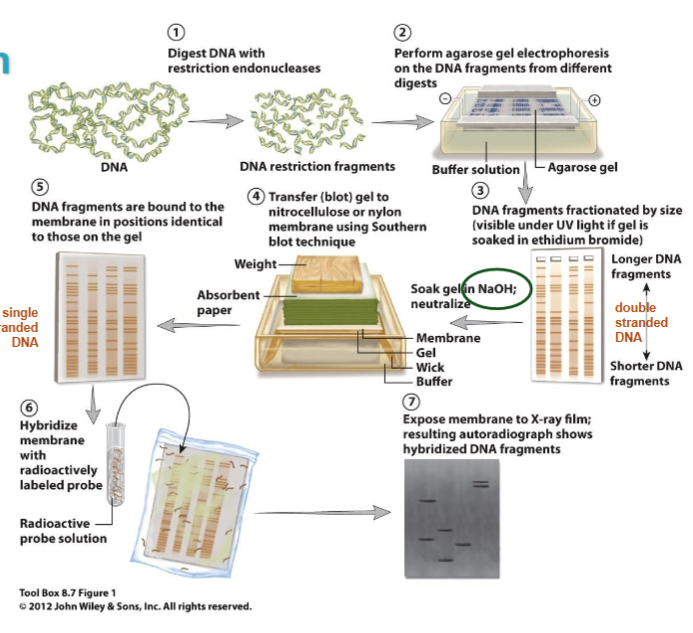
Southern Blot
detects specific DNA fragments by identifying specific restriction fragments in a complex mixture of fragments
can be used for:
estimating # and position of gene copies in a genome
restriction mapping of genomic fragments
detecting cloned sequences, transgenes, homologous sequences in different genomes, repetitive sequences
Gene Expression Studies
DNA is the same in every cell but the proteins made from it is different during the life cycle
temporal control: genes expressed at a precise time during the life cycle of an organism, aka developmental regulation
spatial control: genes expressed in a specific tissue or cell time, aka tissue-specific expression
many genes are temporally and spatially controlled (expressed at a certain time in a certain place during development)
Induced Gene Expression
change in types or amount of gene expression in response to environmental signals, exposure to chemical substance or physiological stress
examples:
thermal stress - heat shock genes
gene expression controlled by steroid hormones
toxins such as heavy metals
antibiotics or anaesthetics
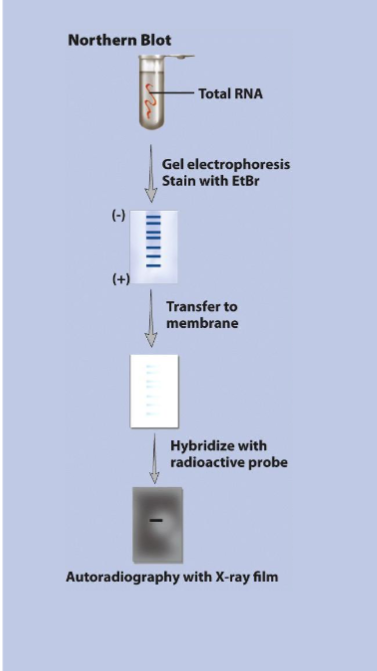
Northern Blooting
detects specific mRNAs
mRNA is transcribed from protein coding DNA (first step in gene expression)
total RNA from all 3 major groups is isolated from cells and electrophoresed
similar to Southern blotting but no denaturation needed since RNA is already ss
can be used to determine stead-state level of a specific transcript in a certain RNA mixture → abundance of specific mRNA at certain time under certain conditions
depends on both transcription and degradation rate for that specific mRNA
Where can we compare abundance of mRNA from?
can compare mRNA isolated from:
different tissues of one organism
same tissue from different organisms
different treatments or conditions
the probe is designed to detect certain mRNA, the one transcribed from target gene and which codes for target protein
In Situ Hybridization
probe binds to complementary nucleic acids in cell or tissue (same probes as for northern or Southern)
difference from northern/Southern is it identifies genes directly in chromosomes and transcripts (mRNA) directly in cell or tissue for developmental expression studies following treatments or environmental changes
FISH
fluorescence in situ hybridization
used to identify genes directly in chromosomes
Expression Studies for Multiple Genes
pattern of genes expressed in a cell is characteristic of its present state
all or most differences in a cell state are correlated with changes in mRNA levels of genes
expression patterns of uncharacterized genes can also give clues about their function
important to study interactions between individual genes → study interaction between genes simultaneously
use microarrays instead of traditional methods
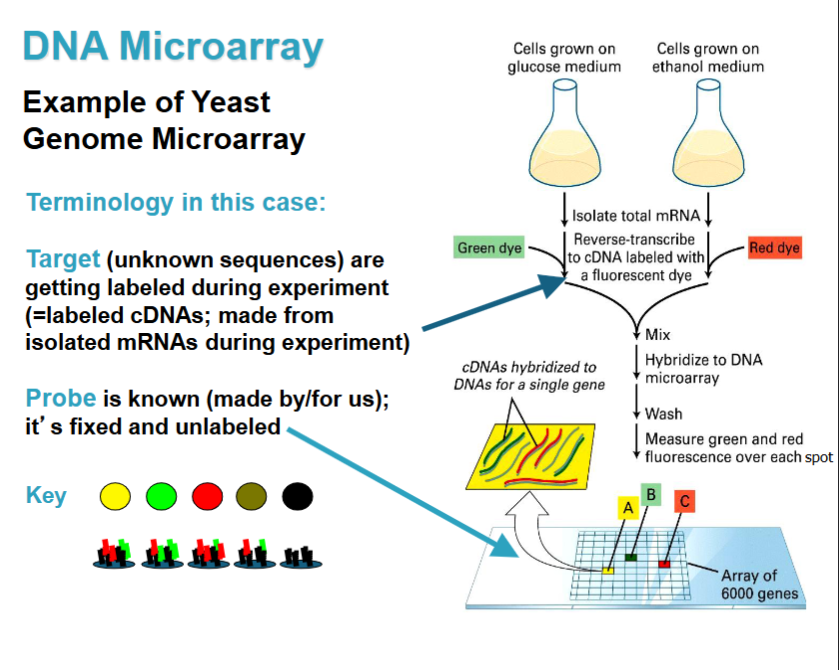
DNA Microarray
target: unknown sequence getting labeled during experiment (cDNAs made from isolated mRNAs during experiment)
probe: what is known for us; fixes and unlabeled
Measuring Gene Expression by Measuring Translation
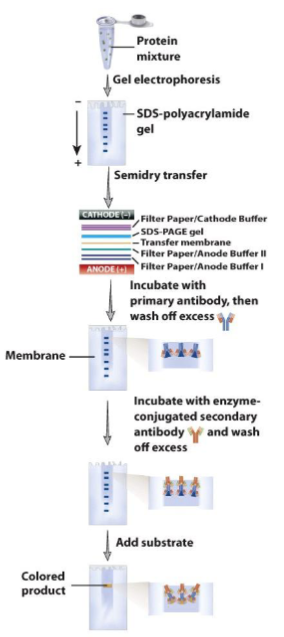
SDS PAGE and western blotting
SDS PAGE: (-) charged detergent to bind to hydrophobic protein regions to help unfold by coating in negative charge that is proportional to their mass
electrophoresis under denaturing conditions → proteins migrate towards (+) electrode when voltage is applied to gel
separate by molecular weight instead of intrinsic charge, detect in gel by different stains
2D gel can be used to isolate by size and charge
Western Blotting
step 1 is SDS-PAGE to separate proteins by size
proteins are transferred to membrane - western blotting
membrane is incubated with antibody-specific for one of the proteins to see if it is expressed in control vs. experimental conditions
bound antibody is detected by secondary antibody that is conjugated to an enzyme or tagged (radioactive or fluo) → visualization