Lecture 6 - Eukaryotic Transcription
1/83
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
84 Terms
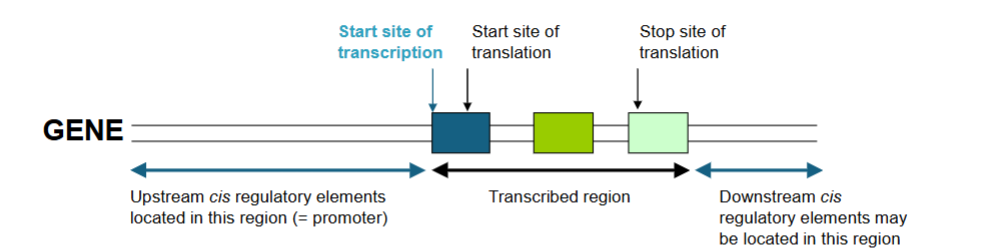
Mapping Transcriptional Start Sites
DNA promoter elements that control transcription are often located near the start site of transcription of the gene and there are important regulatory elements in the 5’ UTR of mRNA
to define the promoter for a gene you need to know the start of transcription
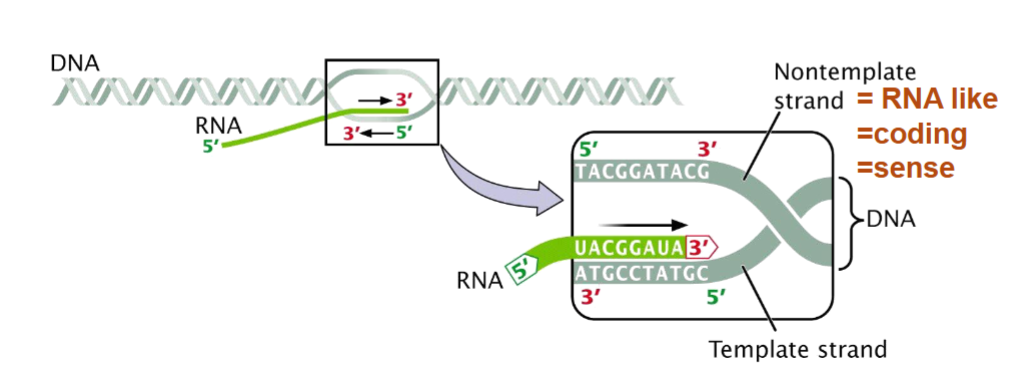
Similarities in Transcription and DNA Synthesis
there is a multi-subunit complex for the enzyme which makes the nucleotide strand in 5’-3’ direction
Mg2+ is a co-factor and is necessary to add to buffers for in vitro transcription
both DNA strands can be templates for RNA synthesis
antisense strand = template
Differences in Transcription and DNA Synthesis
enzyme is RNAP which doesn’t need a primer
promoter sequences where RNAP binds are asymmetrical - RNAP positioned so it can only transcribe one strand from one promoter
DNA is unwound locally only vs energetically favourable state that doesn’t need ATP
product is ssRNA that is released immediately so helix can re-form
precursors use rNTPs
less efficient in proofreading but less important than DNA mistakes since those stay over generations
Gene Expression
process in which information carried by a gene is converted into observable product
Transcription Definition
first step in gene expression where one strand of a DNA molecule is used as a template for synthesis of a complementary RNA (mRNA) which carries info for a specific protein
One-cell System of Gene Expression
in bacteria
cell has to survive and reproduce; gene expression is regulated to adjust to changes in nutritional environment to enable cell growth and division
Multicellular Organism Gene Expression
has to survive and reproduce but also grow and develop; different parts of the body have different functions
gene expression is regulated during:
development (time)
tissue differentiation (space and space/time combo where needs to be on sometimes but not always)
stress (as a response to environmental stress - induction)
Eukaryotic vs. Prokaryotic Gene Expression
prok:
transcription and translation are coupled in the same compartment
mRNA is polycistronic without introns, and has a short half-life
euk:
eukaryotic cell is compartmentalized and there is regulation in each compartment
gene expression can be regulated at various levels
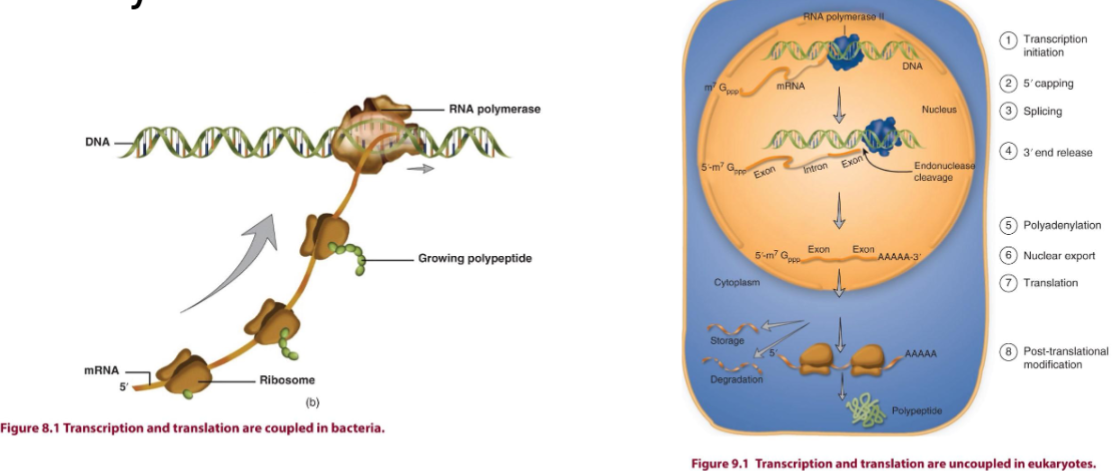
Eukaryotic Transcription
in a different compartment than translation
pre-mRNAs are subject to extensive post-transcriptional mods → processing
chromatic structure limits accessibility (only 0.01% of genes in a typical cell are undergoing transcription at any given moment)
euk RNAP does not recognize binding site by itself → needs general transcription factors (GTF) to help
mostly multicellular organisms with different cells/tissues
3 major RNAPs with different roles
Bacterial RNAP
called RNAP
transcribes all bacterial genes
Eukaryotic RNAPs
RNAP I - transcribes rRNA genes
RNAP II - transcribes mRNA, snoRNAs, some snRNAs, miRNAs
RNAP III - transcribes tRNA, 5S rRNA, some snRNAs
RNAP IV and V (plants only) - transcribes siRNA-directed DNA methylation and gene silencing
mitochondrial RNAP and chloroplast RNAPs also exist
RNAP I Intro
located in nucleolus, where ribosomal genes are concentrated
responsible for transcribing rRNA genes
14 subunits
RNAP II Intro
located in the nucleus
responsible for transcribing the majority of genes: mRNA, snoRNAs, some snRNAs, miRNAs
12 subunits
RNAP III Intro
located in nucleus
responsible for transcribing tRNA, 5S rRNA, some snRNAs
17 subunits
Common Subunits in the RNAPs
rpb 5, 6, 8, 10, and 12
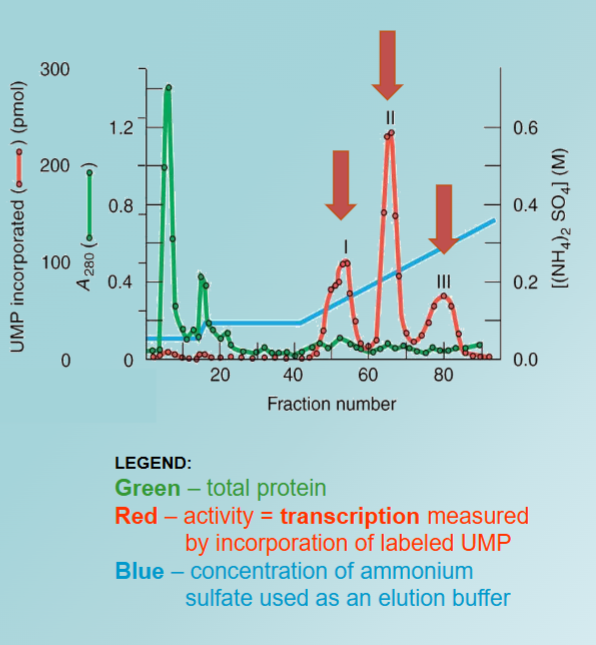
Separation and Purification of RNAPs
(1) rRNA genes:
have high GC content (60%)
are repetitive (up to 20 000 copies of the gene/cell)
found in nucleolus
(2) RNA synthesis:
high ionic concentration - RNA with low GC content
low ionic concentration - RNA with high GC content (similar to rRNA)
Mg2+ low ionic strength - most transcription in nucleolus
Mn2+ high ionic strength - transcription throughout the nucleus
this shows us there is more than one RNAP, one works in the nucleolus, stimulated by low salt and Mg2+, and the other in nucleoplasm stimulated by high salt and Mn2+
Salt concentration in elution buffer will be ____ in each subsequent elution.
higher;
higher salt concentration displaces proteins with positive charge
different fractions will have proteins based on charge differences → use proteins from each fraction for in vitro transcription
this shows clear third RNAP

RNAP I, II, and III and Ionic Strength
RNAP I – active at low ionic strength, works with both Mg2+ and Mn2+
RNAP II – more active at high ionic strength, works better with Mn2+
RNAP III – active over a broad range of ionic strengths, works better with Mn2+
Biochemical Experiments
showed RNAP II is most sensitive to a-amanitin toxin from mushroom since it binds to rpb1 and prevents RNAP II translocation
RNAP I is most sensitive to actinomycin D antibiotic made by Streptomyces since it intercalates into GC rich regions and inhibits transcription
RNAP I
makes rRNA 45S precursor (13 000 nucleotide polymer) → 18S, 5.8S, 28S rRNAs
uses only 1 type of promoter: core promoter + upstream promoter element (UPE) where the efficiency of the core promoter is increased by UPE
upstream binding factor (UBF) and selectivity factor 1 are ancillary factors needed for high-frequency initiation bind to UPE (increases promoter affinity and strength)
RNAP I holoenzyme binds to UBF-SL1 complex at core promoter
2 UBFs bind to minor groove of G:C rich element in UPE and DNA turns to bring UPE and core promoter closer together
lets UBF stimulate binding of SL1 part of a complex → TBP + 3 RNAP I specific TBP-associated factors
TBP is a component of positioning factors needed for initiation by RNAP II and III
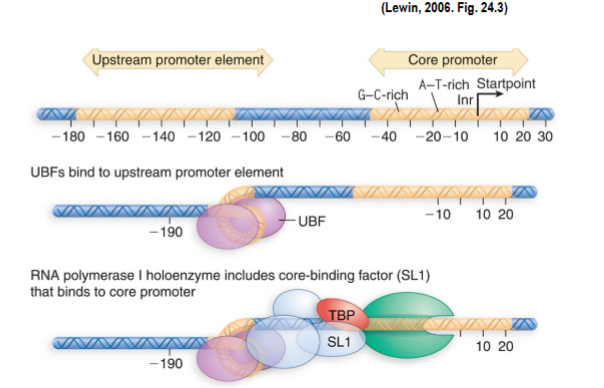
Core Promoter
shortest sequence at which RNAP can initiate transcription typically at low efficiency, interaction with more elements such as UPE increases it
The Nucleolus and RNAP I Transcription
human: clusters of ~40 copies of rRNA genes on 5 chromosome pairs = 400 gene copies
each cluster is a nucleolar organizer region (NOR)
size of nucleolus depends on the metabolic activity of the cell
size means number of clusters involved in rRNA synthesis

RNAP III
tRNAs, other small RNAs including 5S rRNA
has 3 types of promoters recognized in different ways by different groups of transcription factors:
2 types of internal: transcribe 5S rRNA and tRNA genes
upstream: transcribes snRNAs, similar to RNAP II TATA promoters

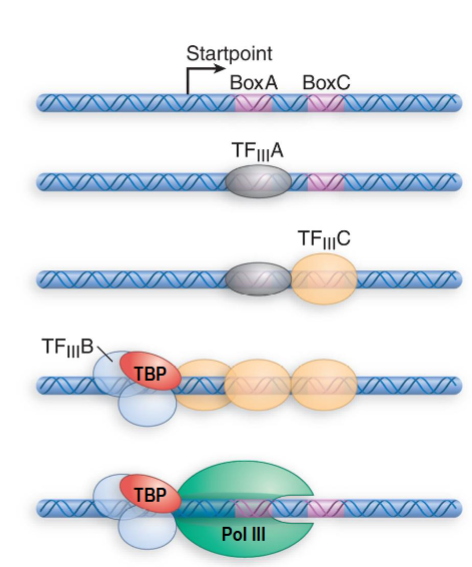
Type 1 RNAP III Promoters
internal type found only in genes for 5S rRNA
TFIIIA to BoxA then TFIIIC binds to BoxC
subsequent binding of TFIIIC displaces TFIIIA and allows TFIIIB (positioning factor) to bind upstream from startpoint
TBP can bind to TFIIIB
RNAP III can be recruited and TFIIIC is displaced
efficiency of transcription is altered by changes in region upstream from the startpoint (spacing or sequence)
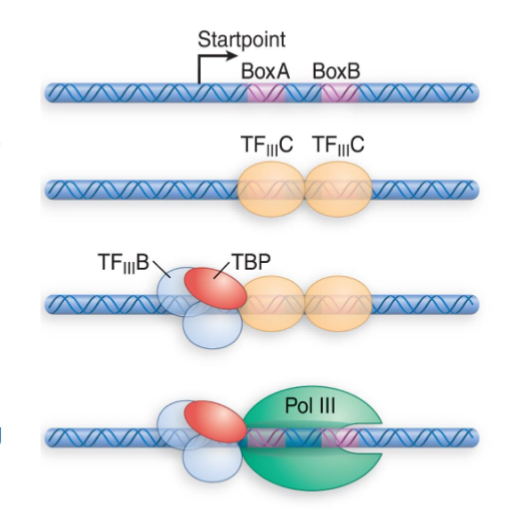
Type 2 RNAP III Promoters
transcribe tRNA genes
TFIIIC binds to BoxA AND BoxB downstream of startpoint
allows TFIIIB (positioning factor) to bind near the startpoint and TBP can be positioned
RNAP III recruited
TFIIIA and TFIIIC are assembly factors to help find TFIIIB
have to be removed without affecting transcription
TFIIIB is the only true initiation factor required by RNAP III
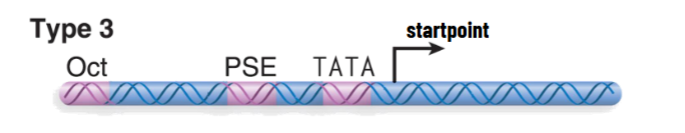
Type 3 RNAP III Promoters
transcribes some snRNAs
more conventional arrangement (like RNAP II) with upstream elements to regulate transcription initiation
TATA element is immediately upstream of startpoint: efficiency increased by other elements - factors binding here act cooperatively
TATA element is bound by TFIIIB that actually recognizes DNA promoter sequence
TFIIIB/TBP for RNAP III has the same role as TFIID/TBP for RNAP II
in all 3 promoter types, TFIIIB binds to promoter to form preinitiation complex → directs binding of RNAP III
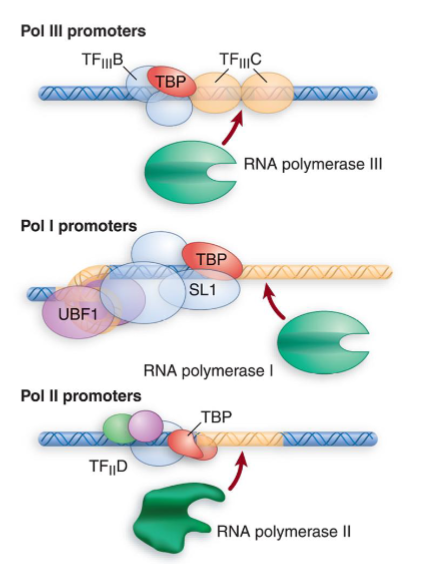
RNAP I, RNAP II, and RNAP III
TATA binding protein is a component of the POSITIONING factor no matter which RNAP is transcribing → allows each type of polymerase to bind to its promoter
involved in coordination of activities of all 3 polymerases through binding to other polymerase-specific factors
TBP = #1 commitment factor that coordinates transcription factors and pulls in the right RNAP
within SL1 complex (RNAP I)
within TFIID (RNAP II)
within TFIIIB (RNAP III)
also in TATA-less II promoters
Structure of RNAP II
x-ray crystallography of 3D yeast RNAP II crystals and posed on synthetic DNA (no promoter)
25 angstrom channel in the face of the RNAP II which can accommodate 20 bp of DNA; channel formed by Rbp1 and Rbp2
at the opening of the channel “jaws” role: grabbing of dsDNA
sliding clamp - composed of parts of Rbp1, 2, 6
active site is on Rbp2 includes Mg2+ and conserved aspartate motif - RNA synthesis (facilitates nucleophilic attack)
two pores - one should be the exit for the growing RNA
basically large subunit structure that make a clamshell that allows DNA and RNA to be transcribed
Core Subunits of RNAP II Structure
rpb1, 2, and 3, are absolutely necessary
rbp1 binds DNA, responsible for a-amanitin sensitivity
2 forms of large subunit: phosphorylation on carboxyl-terminal domain (CTD)
alpha is non-phosphorylated
o is phosphorylated
II alpha containing enzyme binds to promoter; II o-containing enzyme is in elongation phase (functionally different)
rpb2 is the polymerization active site; rpb3 is a 20 aa region of similarity with bacterial subunit a, 2 monomers in holoenzyme
CTD Tail
unique to pol II (not in I or III)
stretch of 7 aas that are repeated multiple times (at least 10) on rpb1 subunit
tyr-ser-pro-thr-ser-pro-ser
5 of these 7 have -OH so this is a hydrophilic, phosphorylation site (mostly polar amino acid residues)
critical for viability
un-phosphorylated CTD tail used to initiate transcription
phosphorylated CTS is present only for high levels of transcription
critical for methyl cap addition and polyadenylation; splicing
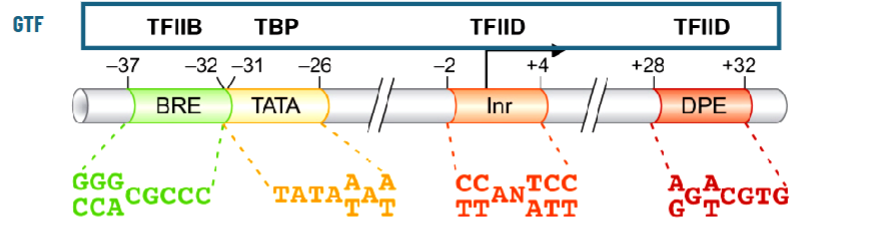
Promoters Recognized by RNAP II
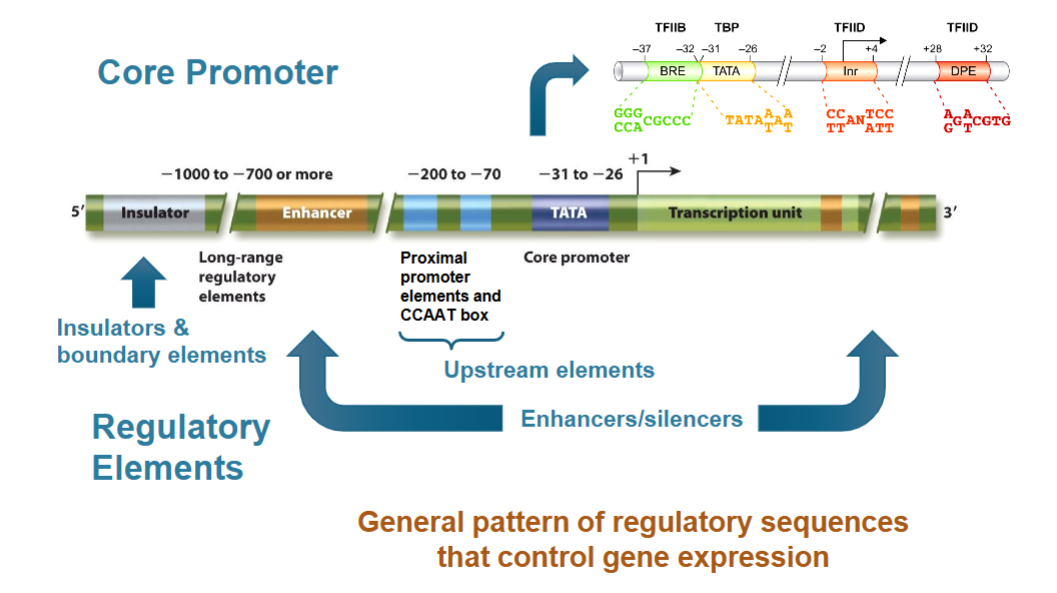
class II promoters (promoter regions) have 2 parts:
core elements (aka core promoter or just promoter)
regulatory elements (one is the enhancer)

Core Promoter of Class II Promoters
minimal set of elements required for accurate in vitro transcription initiation by RNAP II
necessary for recruitment, binding, and proper positioning of RNAP II
TATA box at ~-30 TBP binding site
initiator on TSS
downstream promoter element
TFIIB, upstream
but lots of promoters don’t have initiator and downstream element and there are TATA-less promoters too
Regulatory Elements of Class II Promoters
bind regulatory proteins
classification of the elements (and the regulatory proteins that bind to them) is not always straightforward
presence or absence of regulatory elements (sequences) can influence transcription rates through binding of regulatory proteins
sometimes the regulatory elements are necessary for initiation of transcription
generally classified based on the distance from a core promoter and activity
upstream elements (upstream and relatively close to, but not in, the promoter)
enhancers and silencers (everywhere in any orientation)
boundary elements and insulators
Core Promoter
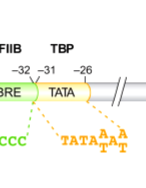
TATA Box:
highly conserved sequence 25-35 bp upstream from start site
similar in action to e. coli TATA box but is further upstream
TATA-less promoters: found in housekeeping genes - absolutely necessary genes or in specialized genes (made in only certain cells)
those genes must have either initiator core element or GC boxes (upstream element) to start transcription

Functions of TATA Box
is an element of the core promoter
SV40 early promoter (viral promoter): WT has 3 possible transcription start sites (all Gs)
deletions of the sequences between TATA and WT start sites indicate transcription will start somewhere downstream of the TATA site → sequence doesn’t matter; DISTANCE from TATA to start is most important
TATA box finds start of transcription ~30 bp downstream
tata box sometimes important for the efficiency of transcription
TBP binds to TATA box and starts assembly of general transcription factors and RNAP
Enhancers/silencers can also be present in ______ ___ or ______.
structural gene; downstream
Initiator Element
part of core promoter; could be part of TATA-less promoters
majority have C at -1 position and A at +1 position
strength of promoter determined by surrounding nucleotide sequence
transcription with initiator elements or TATA boxes begins at precise site
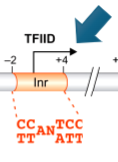
Downstream Elements
part of core promoter; could be part of TATA-less promoters
binds TFIID (general transcription factor)
TFIIB Recognition Element (BRE)
binds TFIIB (General transcription factor)
considered part of a group of regulatory elements
Regulatory Elements
may be present individually or in “sets”
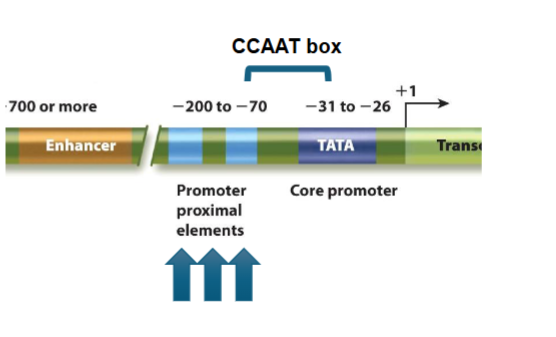
upstream elements:
GC boxes: GC rich stretch, orientation independent (can be flipped 180 degrees) but must be close to TATA box - 20-50 bp upstream of start site, usually in housekeeping genes
could be a part of TATA-less promoter and transcription could be initiated at any one of multiple possible sites over 20-200 bp resulting in mRNAs with multiple alternative 5’ ends (UTRs_
CCAAT boxes: enhancer element 30-75 bp upstream with CTF (CCAAT transcription factor)
no prok. equivalent; may be necessary in euks.
promoter-proximal elements: control regions 100-200 bp upstream from start site with various numbers of sequences and effects
cell type specific - specific set of elements dictates expression
identified through 5’ deletion series
Enhancers
part of regulatory elements
control elements that typically stimulate transcription
could be a multiple binding site for different transcription factors
transcription factors that recognize enhancer = enhancer binding proteins = activators → interact with general transcription factors
sometimes the same element could be both enhancer and silencer based on the protein that is bound
can be thousands of bp away
orientation doesn’t matter
can occur downstream in an intron or in the exon
transvection: enhancers on adjacent chromosomes (local concentration of factors)
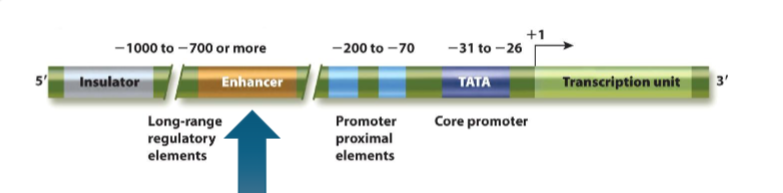
Insulators and Boundary Elements
insulators: form chromatin boundary between euchromatic and heterochromatic regions in an individual chromosome
loops made by CTCF and cohesin, access in under locus control regions’ control
insulated neighbourhoods: regions of DNA within extruded ‘loops’ of eukaryotic DNA; chromosome territories are respected
Review of RNAPII Promoters
class II promoters have two parts: core elements and regulatory elements
core elements include:
TATA box, initiator, downstream element, TFIIB recognition element
regulatory elements include:
upstream elements (GC boxes, CCAAT boxes, promoter proximal elements)
enhancers and silencers
boundary elements and insulators
How do we recognize the RNAP II promoter?
position RNAP II at transcription initiation sites and bend DNA (promote tight binding) to prevent nucleosome reformation
required by transcription of most genes that are transcribed by RNAP II
transcription initiation complex
TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH where D is the largest and is made of TATA binding protein and TBP-associated factors
has 2 basic roles: foundation for transcriptional complex and prevent nucleosome stabilization in the promoter region
The transcription initiation complex is made of _____ and _____.
RNAP II; GTF bound to promoter region
Importance of TBP
it is the first protein to bind to the TATA box → is a positioning factor
it is highly conserved with similar C-terminal domains
monomer that makes a saddle shape and bends minor groove
alternative TBP in small # of cells (TRF1 is very rare and cell-specific)
positioning factor so helps RNAP find its promoter, no matter which is transcribing
within SL1 complex (RNAP I)
within TFIID (RNAP II)
within TFIIIB (RNAP III)
Also in TATA-less II promoters
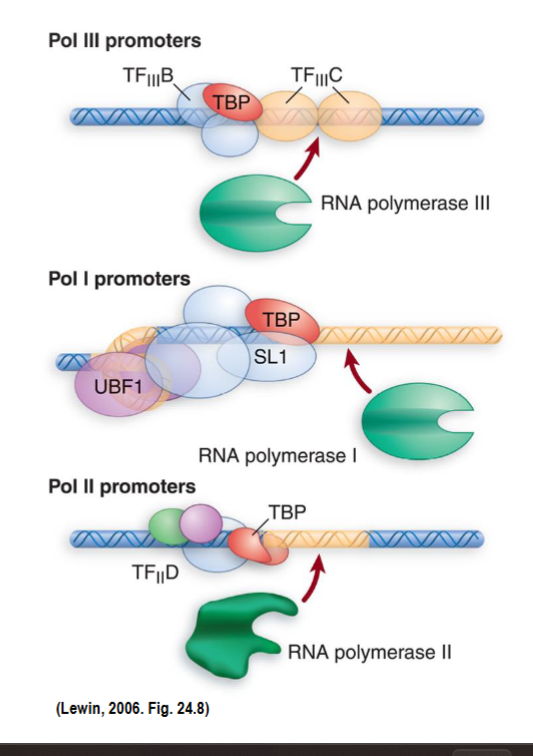
Which factor determined whether or not TFIID stays at the promoter?
TAFs - TATA-associated factors
some promoters are more difficult to transcribe from than others
Steps of Eukaryotic Transcription
TFIID binds at TATA via TBP and TAFs
once TBP/TFIID is bound, TFIIB (monomeric protein) binds - rate limiting step
TFIIF binds to mediator RNAP II → make a preformed complex and direct mediator RNAP II to GTFS assembled on the promoter
2 large RNAP II subunits interact with TFIIB which is already positioned on DNA
unphosphorylated CTD tail of RNAP II is in direct contact with TFIID
TFIIE and TFIIH bind to the complex (TFIIE = DNA-dependent ATPase, TFIIH = helicase and protein kinase activity)
TFIIH helicase activity unwinds DNA with energy from TFIIE, kinase activity phosphorylates CTD tail of RNAP II
phosphorylation of CTD tail detaches RNAP II from TFIID and promoter
transcription pause released by more phosphorylation of CTD and other factors by CDK9
promoter clearance (mediator and GTFs released)
RNAP II goes to elongation
Which TF is critical for RNAPII orientation on the promoter?
TFIIB; near RNA exit site and active site
Promoters Lacking TATA
in majority, TBP needs to be placed at the right spot first with respect to existing core element and transcription start site
initiator element:
TBP can bind to promoter itself with initiator minus TATA through TBP-associated factors (TAFs) → TFIID binds
GC boxes:
TBP can’t bind itself to promoter with GC but no TATA through TAFs → TFIID binds
after TFIID binding, events are the same as in TATA promoters
Proteins that Affect Transcription
basal transcription is seen in vitro; in vivo, euk promoters have to be activated by upstream bound transcription factors
GTFs and TFs directly influence RNAP binding:
GTFs bind to core promoter or RNAP II or each other to anchor RNAP II
TF (activators and repressors) bind to DNA at proximal elements, enhancers/silencers, even core promoter elements
co-activators and mediators bind to both activators and basal apparatus as a bridge
chromatin remodelling enzymes and other regulators act on local chromatin structure
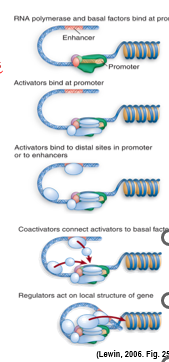
In order for a protein to be a transcription factor it:
has to be able to bind DNA
has to be able to activate or repress transcription
transcription factors are modular proteins → different domains have different functions
DNA-binding domains (DBD) interacts with DNA sequences to bind DNA
transcription activation domain (AD) interacts with other proteins to stimulate or impair transcription from a nearby promoter
some have dimerization domains for homo or hetero dimers
ligand-binding domains bind small molecules to regulate TF activity
Domains vs. Motifs
domains: tertiary structure of large proteins organized in distinct regions of the proteins; can be active and stable on its own when removed from the rest of the protein
motifs: specific combinations of secondary structures which are organized into specific 3D structure inside the domains (but refer to protein secondary structure)
DNA Binding Domain Overview
Zn fingers (C2H2, C4, C6)
one alpha-helix in homeodomain
basic alpha-helix part in bZip
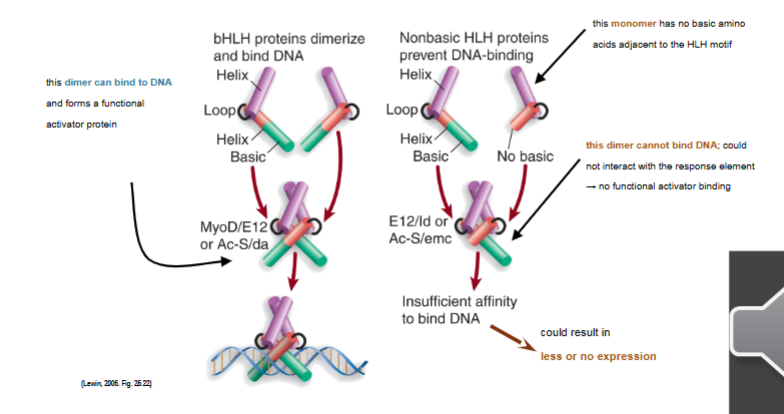
basic alpha-helix in bHLH
DNA Binding Domain
structural motif present in euk transcription factors responsible for its interaction with specific DNA sequences
has a variety of structural motifs for the classification of transcription factors based on interactions and how they bind with DNA
Structural Motifs in DNA Binding Domain
zinc finger: proteins with regions around a zinc ion also seen in protein that do not bind to DNA, 3 types; only one that can bind as a monomer
homeodomain protein: one of the oldest forms of DBD and are important in development of organisms, a highly conserved 60 aa sequence organized into 3 alpha-helices, two of which are organized into helix-turn-helix and third fits into major groove with N-terminal into minor groove
leucine-zipper proteins (bZIP) have hydrophobic leucines in every 7th position of the C-terminal end of the protein (every second turn) for dimerization
helix-loop-helix (HLH) is similar to zipper motif with hydrophobic residues on one side of C-terminal and aloha helix dimerization; tend to form dimers with the second alpha helix responsible for DNA binding to make homo or heterodimers
Types of Zinc Fingers
C2H2 holds zinc with 2 Cys and 2 His residues, alpha helix inserts into major groove of DNA - usually 3 or 4 finger domains, can bind by self as a monomer
C4 has 2 groups of 4 Cys residues binding 2 zinc ions binding as a homo or heterodimer (homo needs 2 copies, hetero needs another copy of a different protein to bind)
C6 zinc finger in yeast uses 6 Cys residues to bind 2 zinc ions into a globular domain and small recognition helix; requires alpha helix dimerization domain (homo or hetero doesn’t matter just needs dimerization)
Transcription Activation Domain Overview
acidic
glutamine
proline rich
Transcription Activation Domains
allows for proteins to do protein-protein interactions for activation/repression of transcription
protein-protein interactions allow for conformational changes in the protein for it to activate or repress
acidic activation domains interact with co-activator
some yeast activators, mammalian glucocorticoid receptor, herpes virus activator
lots of aspartic and glutamic acid
glutamine-rich domains
proline-rich domains
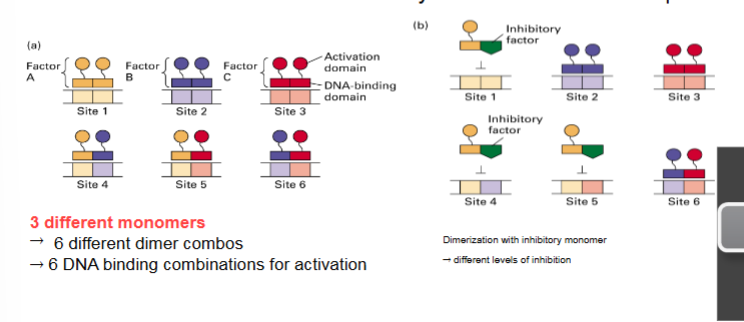
DNA Binding and Transcription Activation Domains
experiments with chimeric hybrid proteins show activating domain and DNA binding domain are separate and independent
Gal4 is an activator with DNA binding and activation domain and Lex A is a repressor with DNA binding and activating domain that acts as a repressor → cut off repressing activating domain of Lex A and hook up to activation domain of Gal4 → whole protein acts as an activator when bound to lexA sites in DNA
DNA binding domain from repressor + activation domain from activator = acting as activator even though DBD is from a repressor
therefore, activation domain is important ONLY for activation and DNA binding domain is ONLY for binding; they are independent
Dimerization Domain Overview
one of the Zn fingers (alpha helix in zinc finger C6)
leucine zipper part in bZip
second helix in bHLH
Ligand Binding Domain Overview
in Zn finger - C4 factors where the steroid hormone is the ligand
Dimerization Domain
allows for greater diversity and complexity of factors (more control of gene expression)
combinatorial control of different activation domains into a third activation domain
recognition of different binding sites
heterodimerization can alter DNA binding specificity of transcription factor for combinatorial control → control by combination of different proteins
dimerization does robust activation compared to monomer itself since it can recognize more sites
Ligand Binding Domain
a ligand is any type of effector molecule or covalent attachment
binding/modification of this domain will cause conformational change and modulate the native active of the regulatory molecule
is typically reversible and there are a variety of ligands (e.g. end products like tryptophan, carbon sources like lactose, covalent modification like phosphorylation)
example: CCAAT Enhancer Binding Protein B is a transcription factor for endometrial cells that can be phosphorylated to become active and open up → big structural change
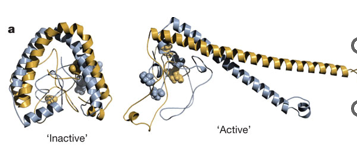
Enhancers
any TFs that bind can interact
have multiple binding sites as landing pads for transcription factors
cooperative binding makes an enhanceosome, a nucleoprotein complex by bending or looping DNA
binding certain proteins makes assembly of other proteins → combinatorial control
gene is only expressed with the correct combination of TFs in the cell
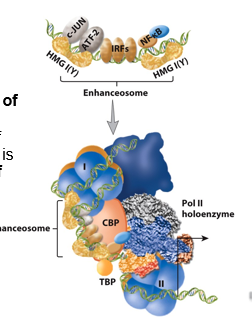
Co-activtors
make direct contact with activators or repressors through transcription activation domain
are not transcription factors themselves so need to bind something else to regulate transcription
have specific functions, such as histone modification of tails to affect the degree of compaction of nucleosome → allow for greater accessibility of TFs to the transcribed gene
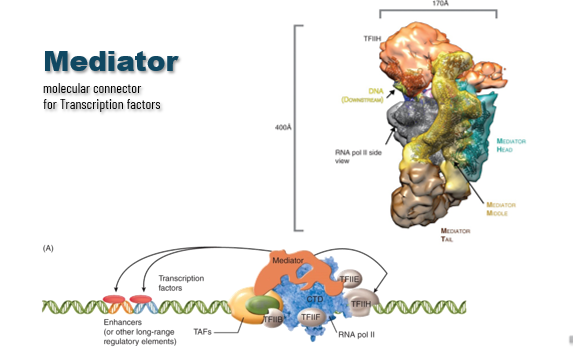
Mediator Complex
protein complex needed for assembly of the pre-initiation complex (RNAP II and GTFs) and successful initiation
allows for fine-tuning and cooperation of proteins (Remodeling of local region of transcription) → does this through activator domains contacting different mediator components
function is to coordinate a combination of activators/elements and can do conformational changes of the complex → changes rate of RNAP II initiation (engages more quickly or more slowly)
helps RNAP II coordinate the signals it’s getting from all the TFs
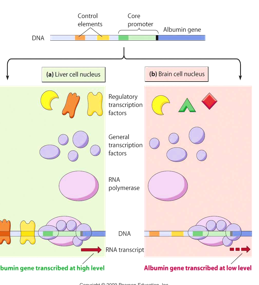
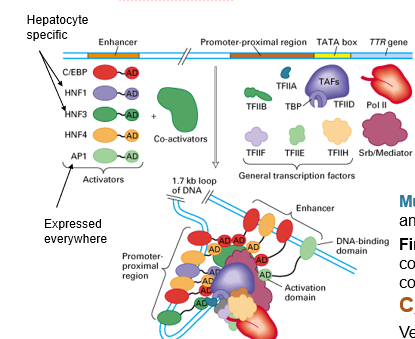
Tissue Specificity of Cis Regulatory Elements
DNA is the same in every cell/tissue therefore the cis elements are the same everywhere but we don’t want all the same proteins expressed everywhere
tissue specificity comes from the action of tissue-specific TFs
regulatory proteins are tissue specific and this is what dictates the tissue specificity of cis elements (crucial for transcription initiation)
Transcription Initiation by RNAP II
highly regulated in proks and euks because it has to respond to developmental timing and environmental changes
one gene can be transcribed in multiple condiitons
different conditions are associated with different cis elements which bind different transcription factors
combinatorial control since different factors at each site will interact and that determines if the gene is turned on and expressed
no consistent location for where cis regulatory elements will be found - a factor’s presence/absence could increase/decrease transcription
different genes that respond to the same signal in the same way all have the same cis element
promoters are modular - variety of elements can contribute to promoter function meaning none are essential or common to all promoters
elements can differ in number, location, and orientation
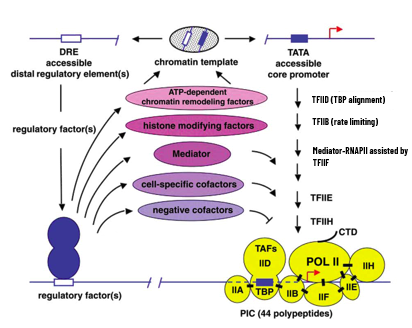
Influences on Transcription Initiation
direct influence on TFs on assembly of initiation complexes
recruitment of GTFs
co-activators
interactions
enhanceosomes
architectural changes
TFs regulate changes in chromatin structure (remodeling) and control histone acetyl and deacetylation
concentration and activities of TFs
protection of active gene promoters from methylation
Recruitment of GTFs
RNAP in euks is not directly recruited to DNA unlike proks that can scan and find
TFs help recruit GTFs (which associate with RNAP)
holoenzyme model: RNAP II and GTFs are recruited as a preassembled whole that lands on the transcription unit
enhanceosome model: happens stepwise where TFIID complex binds first due to interactions of TAFs with activators bound to the enhancers (opens up region to get access, TFs assemble) but steps may happen in parallel
Co-activators
interact with TFs with modulator complex, co-activators, each other, ligands → promote or prevent GTF binding
e.g. CREB protein that binds cAMP response element and activate associated genes by binding to CREB-binding protein (CBP) → CREB is activated by phosphorylation to allow association with cAMP response element (CRE)
Interactions
TFs interact with each other or ligands, those interactions promote or prevent binding of GTFs
Interactions Involving Repressor Proteins:
repressor inhibits gene activation by binding to site that overlaps activator site
repressor binds adjacent to activator and interferes with AD (activation domain) of activator
repressor binds upstream and interacts with mediator through GTFs to inhibit initiation
co-repressors recruited that alter nucleosomes to inhibit transcription
Interactions Involving Ligand Binding Domains
both activation and inhibition switch on and off depending on if ligand is bound or not due to conformational changes in the TF
retinoic acid
Interactions Involving Enhanceosomes
TFs make these at any distance from GTF binding site for specific gene expression, usually for tissue-specific gene expression
multiple enhancers and modular arrangements give fine control through different combinations and concentrations of TF → combinatorial control
Interactions Involving Architectural Changes
TFs bind to specific binding sites to change the shape of the DNA in a control region → changing interactions transcription factors, and/or GTFs, stimulating transcription
difficult for DNA to bend or loop if the distance between core promoter and enhancer is too short, may need more architectural TFs
Interactions Involving Insulators
sets up boundaries between DNA domains - prevents activation or repression of genes by close but unrelated activators and repressors

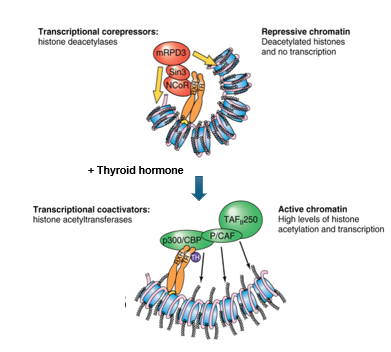
Histone Acetylation and Deacetylation
TFs regulate this
core nucleosome has histones with a flexible tail where modifications occur to compact or loosen the chromatin
20-40 aa N-terminal tails have positive lysine residues that can be acetylated or deacetylated
acetylated = positive charge is neutralized and eliminates interaction with DNA → DNA less condensed, promoter regions available (euchromatin)
catalyzed by histone acetyltransferases (HAT) associated with activation of gene expression
deacetylation → repression of gene activity where deacetylases are in co-repressor complexes
Histone acetylation is ____ but not ____ for activation.
essential; sufficient
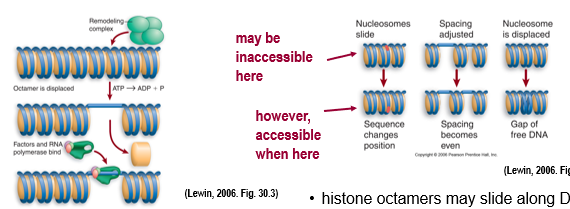
acetylation of histone tail makes the chromatin less compact but we still have nucleosomes intact which need to be repositioned to expose promoter elements - chromatin remodeling protein complexes
use ATP energy to remodel and move nucleosomes out of the way (displace histone or slide it along DNA)
swi/snf proteins are important for altering structure of nucleosome core, swi proteins move/remove/replace nucleosomes on DNA
remodeling complexes don’t bind DNA sequences themselves → are recruited by activators or repressors
Concentrations and Activities of TFs
regulated during cell differentiation and in response to hormones and signals from other cells
as we have differences in concentration, we make a gradient that affects which genes will be expressed (think Drosophila local concentration of TFs in different parts of the organism that affects different genes
critical point: transcription of genes which are transcription factors themselves
transcription activation based on concentration of effected cell important during development
accessible binding sites and respective binding proteins are necessary for transcription
Protection of Active Gene Promoters
protecting from methylation
some promoters have CpG islands are preserved as unmethylated to keep genes active
islands are stretches of 200-500 nucleotides with GC content more than 50% that act as core promoters or upstream elements
mechanism of protection is still unclear but could be prevention of DNA methyltransferase binding, demethylase existing, GTFs and RNAP II and H3 tail mods excluding DNA methyltransferase (DNMT) from sites of transcription initiation
could also reduce amount of GC pairs but we can’t control the DNA so we just don’t have CpG islands in some areas
deamination of normal C to U or methylated C to T which gets recognized by repair system and get fixed as a mutation (getting rid of methylated C through conversion to T)

Which enzyme recognizes single strand methylation and repairs the new strand?
maintenance methylase; there are no methyl groups on newly made DNA strand, the parent strand is methylated
involved in protecting active gene promoters from methylation
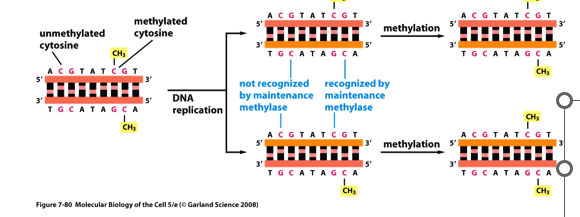
CpG methylation is known as _____ _____.
epigenetic modification; means on top of the gene
the DNA sequence is not changed, just methylated which changes gene expression
if methyl groups are removed from CpGs in promoter regions, transcription is able to occur but not necessarily will
no methylation = necessary but not sufficient for transcription to occur but this is not a universal method for regulation of gene expression (e.g. drosophila)