BIOL 141 POLL EXAM 3
1/90
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
91 Terms
Mitosis is responsible for what key processes in eukaryotes?
Wound repair
Growth
Asexual reproduction
All of the above
All of the above
In eukaryotic cells, chromosomes are composed of
DNA only
RNA only
Protein only
DNA & RNA
DNA & protein
RNA & protein
DNA & protein
How many DNA double helices are there in an unreplicated chromosome?
0.5
1
2
3
4
1
How many DNA double helices are there in a replicated chromosome?
0.5
1
2
3
4
2
How many DNA double helices are there in a sister chromatid?
1
At what stage of the cell cycle is DNA replicated?
GO
G1
S
G2
M
S
If a cell went through repeated cell cycles consisting of S and M phases only, what would be the most obvious outcome?
Cells would not be able to replicate their DNA.
Daughter cells would have twice the DNA of the parent cell.
Cells would get larger and larger.
Daughter cells would have half the DNA of the parent cell.
Cells would get smaller and smaller.
Cells would get smaller and smaller.
When does the spindle apparatus form?
anaphase
cytokinesis
G1
metaphase
prophase
S phase
telophase
prophase
If a cell completed mitosis but not cytokinesis, the result would be a cell with
A single large nucleus with twice the normal amount of DNA
High concentrations of actin and myosin
Two nuclei, each with the normal amount of DNA
Two nuclei, each with half the normal amount of DNA
Two nuclei, each with the normal amount of DNA
Scientists isolate cells in various phases of the cell cycle. They find a group of cells that have 1.5 times the DNA relative to G1 phase cells. The cells of this group are
between the G1 and S phases in the cell cycle
in the S phase of the cell cycle
in the G2 phase of the cell cycle
in the M phase of the cell cycle
Future investments
in the S phase of the cell cycle
Which cellular event initially commits a cell to cell division?
Entrance into GO
DNA replication
Chromosome condensation
Separation of sister chromatids
Cytokinesis
DNA replication
Cyclin-dependent kinase is
inactive in the presence of cyclin
an enzyme that attaches phosphate groups to other proteins
present only during the S phase of the cell cycle
A and B
Band C
A and C
all of the above
B
What is the consequence of missing or defective Rb (retinoblastoma) proteins for cell-cycle regulation?
G1 cyclin is overproduced, resulting in unregulated progression past the G1 checkpoint.
E2F is active in the absence of G1 cyclin, resulting in unregulated progression past the G1 checkpoint.
E2F is inactive in the absence of functional Rb, resulting in unregulated progression past the G1 checkpoint.
E2F is active in the absence of MPF cyclin, resulting in unregulated progression past the G2 checkpoint.
E2F is active in the absence of G1 cyclin, resulting in unregulated progression past the G1 checkpoint.
MPF, or mitosis-promoting factor, consists of two important cell-cycle regulatory proteins called
actin and myosin
rubisco and ATP synthase
tubulin and kinesin
cyclin and cyclin-dependent kinase
cyclin and cyclin-dependent kinase
The product of the p53 gene…
inhibits the cell cycle
slows down the rate of DNA replication by interfering with the binding of DNA polymerase
causes cells to reduce expression of genes involved in
DNA repair
allows cells to pass on mutations due to DNA damage
Inhibits the cell cycle
The MPF complex turns itself off by
binding to chromosomes
exiting the cell via carrier proteins
activating a process that destroys cyclin proteins
activating an enzyme that destroys the Cdk proteins
activating a process that destroys cyclin proteins
Which of the following are tumor suppressors?
MPF
p53
Rb
MPF & p53
MPF & Rb
p53 & Rb
all of the above
P53 and Rb
A cancer cell would have:
Defects in multiple cell cycle regulatory proteins
Homologous chromosomes...
carry the same alleles
carry information for the same traits
are identical
align on the metaphase plate in meiosis Il
carry information for the same traits
Tumor suppressor genes:
are frequently overexpressed in cancerous cells
encode proteins that help prevent uncontrolled cell growth
are cancer-causing genes introduced into cells by viruses
often encode proteins that stimulate the cell cycle
encode proteins that help prevent uncontrolled cell growth
A species has eight distinct types of chromosomes. A haploid cell of this species contains how many chromosomes?
4
8
16
32
It is impossible to determine based on the information given.
8
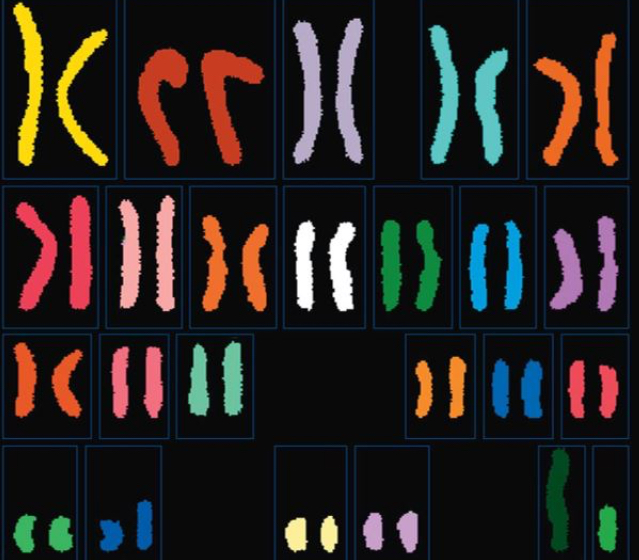
The figure below is a karyotype showing chromosomes from a human cell. What is the haploid number (n) for humans?
23
If gametes were produced by mitosis instead of meiosis with no other changes in the animals life cycle, chromosome number would:
remain the same from one generation to the next
double each generation
be half that of the previous generation
Double each generation. When we think of the life cycle, meiosis is important to REDUCE chromosome number so we can maintain chromosome number from one generation to next. We use the process of fertilization to reduce diploid chromosome numbers. Meiosis halves the number, fertilization brings it back to 2n.
What is the fundamental difference between mitosis and meiosis?
Mitosis involves two cell divisions, whereas meiosis only involves one.
The number of DNA molecules per cell nucleus is cut in half in meiosis but remains constant in
mitosis.
Meiosis occurs in prokaryotes, while mitosis occurs in eukaryotes.
The number of chromosomes doubles in meiosis, whereas it stays the same in mitosis.
The number of DNA molecules per cell nucleus is cut in half in meiosis but remains constant in mitosis.
In haploid, the daughter cell should have half the amount of DNA and chromosomes compared to parents. The goal of mitosis is IDENTICAL cells. Changing chromosome number in mitosis is BAD. Goal of meiosis IS to have DIFFERENCE, reduce chromosome number and genetically distinct daughter cells
Some viruses consist only of a protein coat surrounding a nucleic acid core. If you wanted to radioactively label the protein but not the nucleic acid, you would use radioactive
A. phosphorus
B. sulfur
C. nitrogen
D. carbon
A or C
B. Sulfur
Which of the following would be a prediction of the protein hypothesis?
A. Radioactive protein will be in the pellet.
B. Radioactive DNA will be in the pellet.
C. Radioactive protein will be in the solution.
D. Radioactive DNA will be in the solution.
A&D
B& C
A and D
Genetic information is passed from generation to generation in the form of
Chromosomes
When does DNA replication take place regarding meiosis?
DNA replication does not take place in cells destined to undergo meiosis
before meiosis I begins
during prophase I
between meiosis I and meiosis Il
during prophase Il
twice - once before meiosis I and again between meiosis I and I|
Before meiosis I begins, same like mitosis. Meiosis, we separate chromosomes to produce nonidentical daughter cells, but DNA replication happens BEFORE dealing with chromosomes.
In meiosis 1 we separate homologous chromosomes. At the end, each daughter cell has 1 COPY of EACH TYPE of chromosome, each daughter cell is haploid. In meiosis II, we take haploid cell and reduce DNA BY HALF. We take it from replicated to unreplicated chromosomes. At end of meiosis 1 we have haploid cells but too much DNA.
Which of the following statements is/are true? (Mitosis VS meiosis)
A In both mitosis and meiosis I, homologous chromosomes move independently of each other.
B Meiosis II is similar to mitosis in that sister
chromatids of each chromosome separate.
C In meiosis Il as well as in mitosis, a diploid cell! divides.
D Two of the 4 daughter cells produced at the end of meiosis are genetically identical to the parent cell.
B and C
Meiosis II is similar to mitosis in that sister chromatids of each chromosome separate. When do homologous chromosomes move independently of each other? MITOSIS! Each chromosome is handled independently.
Sister chromatids separate during meiosis and become independent chromosomes. Are the two chromatids genetically identical?
No, because each chromatid came from a different parent.
No, because they crossed over with non-sister chromatids during prophase I.
Yes, because neither of the sister chromatids crossed over during prophase ll.
Yes, because identical chromatids result when chromosomes replicate during S-phase.
No, because they crossed over with non-sister chromatids during prophase I.
In late prophase 1 we have physical contact, regions where they come together and start to exchange pieces. PARTS of chromosome will base pair with homolog. Cross over physically and exchange of pieces. End up with new mixes of pieces.
In the absence of crossing over, will the process of meiosis 1 result in the formation of two, genetically identical daughter cells?
Yes, because each daughter cell contains the same number of chromosomes.
Yes, because identical copies of parent cell DNA are made in S phase, and each copy ends up in a daughter cell.
Yes, because crossing over during prophase I is what produces a unique mix of paternal and maternal alleles.
No, because each daughter cell contains a different mix of paternal and maternal chromosomes.
No, because each daughter cell contains a different mix of paternal and maternal chromosomes.
Crossing over is a mechanism that gives genetic variation, but it’s not the ONLY method. Independent assortment! Each set of homologs move independently. In metaphase I where they line up and daughter cells get 1, each gets a random dad or moms. During metaphase, paternal and maternal line up randomly. When they line up, you have 1 mom, 1 dad, etc. In the next round of meiosis, we end up with either daughter cells have two copies of paternal and maternal, or 1 dad and 1 mom.
We have 23 chromosome pairs, there’s a lot of different ways ours can line up.
Self-fertilization produces offspring genetically identical to parent. True or false?
False
What makes daughter cells different in daughter cells in meiosis?
changing environment
genetic recombination during mitosis
genetic recombination during meiosis
purifying selection
Sexual reproduction does not produce genetically different offspring.
genetic recombination during meiosis
If both homologs in meiosis I move to the same pole of the cell, what will be the “ploidy” of the gametes?
All gametes will have 2n chromosomes.
All gametes will have n chromosomes.
Two gametes will have n+1 chromosomes, and two will have n-1 chromosomes.
Two gametes will have n chromosomes, one will have n+1 chromosomes, and one will have n-1 chromosomes.
Two gametes will have n+1 chromosomes, and two will have n-1 chromosomes.
If both chromatids in meiosis II move to the same pole in one of the two dividing cells, what will be the “ploidy” of the gametes?
One will end up with two chromatids.
In an analysis of the nucleotide composition of DNA, which of the following will be found?
A % = C %
G % + C % = T % + A %
A% = G% and C% = T%
A% + C% = G% + T%
None of the above
A% + C% = G% + T%
When is DNA replicated?
prophase
metaphase
anaphase
telophase
interphase
Interphase.
The leading strand is synthesized ______ and the lagging strand is synthesized ______.
5 to 3, 5 to 3
What holds the strands of a DNA double helix together?
A. Covalent bonds
B. Disulfide bonds
C. Hydrogen bonds
D. Phosphodiester bonds
E. Proteins
A and C
C. Hydrogen bonds
_____ starts DNA synthesis at the 3’ end of the primer.
DNA ligase
DNA polymerase I
DNA polymerase III
Helicase
Primase
Single-strand binding protein
DNA polymerase III

Assume the following DNA sequence with a 6 base RNA primer. What new DNA sequence would DNA polymerase add to the bottom strand?
5 to 3, left to right. C is the answer, look to the RIGHT of the RNA primer.
The leading strand and the lagging strand differ in that…
A. The leading strand is synthesized in the same direction as the movement of the replication fork, and the lagging strand is synthesized in the opposite direction.
B. The leading strand is synthesized by adding nucleotides to the 3' hydroxyl (OH) end of the growing strand, and the lagging strand is synthesized by adding nucleotides to the 5' phosphate (PO4) end.
C. The leading strand is synthesized continuously, whereas the lagging strand is synthesized in short fragments that are ultimately stitched together.
A and B
A and C
B and C
C. The leading strand is synthesized continuously, whereas the lagging strand is synthesized in short fragments that are ultimately stitched together.
_______ removes RNA primers and replaces them with DNA nucleotides.
DNA ligase
DNA polymerase !
DNA polymerase III
Helicase
Primase
Single-strand binding protein
DNA polymerase I comes in. Attaches to 3’ end of fragment, that gives it a place to add a nucleotide. It will pull out the RNA nucleotides that the primase put in on the ADJACENT fragment. DNA polymerase 1 extends from 3’ end, removes RNA and adds DNA. Eventually we end up with a sequence that doesn’t have any RNA in it. We now need to connect the fragments using LIGASE.
During DNA replication the lagging strand is _____
A. Synthesized continuously
B. Made up of short DNA/RNA fragments
C. Synthesized in the 3' to 5' direction
D. Made up of only DNA during replication
A&D
B&C
A & C
B
A student mixes various molecules needed for DNA replication. Replication occurs when he adds DNA, but each duplex has a normal DNA strand paired with numerous DNA fragments <1000 bases long. What has been left out of the mixture?
ATP
DNA polymerase I
DNA polymerase III
DNA ligase
NTPS
Primase
DNA LIGASE. If we have gaps that means we didn’t join together fragments. LIGASE is an enzyme that is defective here. ATP is Adenosine triphosphate. We need dNTP, NOT ATP. DEOXYribonucleotide and a plain old ribonucleotides.
The human genome consists of 3 billion base pairs of DNA. DNA replication is highly accurate. It results in one mistake per billion nucleotides. For the human genome, how often would errors occur, on average?
Six times every cell division. Do the math.
3×10^9 base pairs is size of genome.
1 mistake/1×10^9 nucleotides * 2 nucleotides per base pairs. Nucleotides cancel! Do the math and you get SIX TIMES EVERY CELL DIVISION. BASE BAIRS AND NUCLEOTIDES ARE NOT THE SAME THING.
There are 2 nucleotides in a base pair.
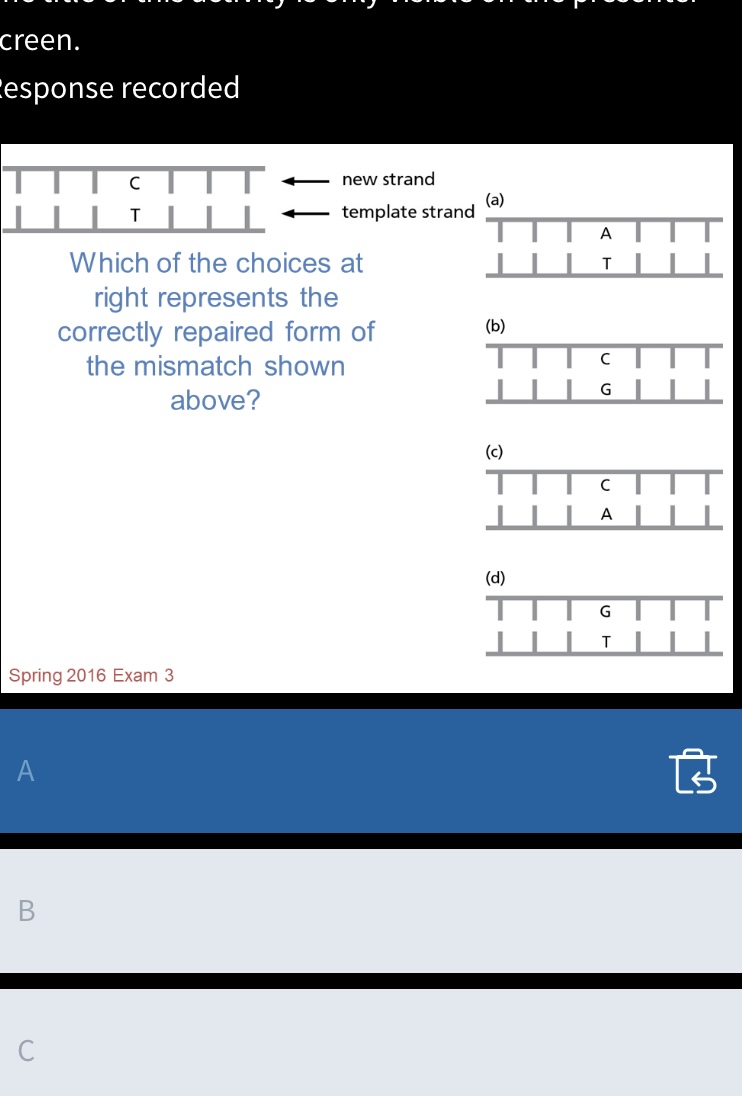
A
Nucleotide excision repair:
removes nucleotides that are incorrectly
incorporated during DNA synthesis
corrects errors in nucleotide excision
recognizes and removes mismatched bases after DNA synthesis is completed
recognizes and repairs thymine dimers and other damaged bases in DNA
recognizes and repairs thymine dimers and other damaged bases in DNA. This is NOT related to DNA replication and synthesis. This is NOT mismatch repair, this is repairing DAMAGED DNA. When DNA is damaged we have to fix it because it can’t base pair in the normal way. UV light hits skin and it causes a new set of covalent bonds to form between TWO THYMINES in a row. Changes shape.
What aspect of DNA structure makes it possible for the enzymes of nucleotide excision repair to recognize many different types of DNA damage?
the energy differences between correct and incorrect base pairs
the antiparallel orientation of strands in the double helix
the regularity of DNA's overall structure
the polarity of each DNA strand
The regularity of DNA’s overall structure. When we think about DNA structure, it has a double helix secondary structure. Spacing is the SAME. A and T and G and C have same spacing. Enzymes can scan along and see if there’s same different. Thymine dimmers cause diff spacing.
In a cell, a gene is…
all the hereditary material in the cell
B. transcribed once per cell cycle
C. a section of DNA that contains the regulatory sequences and coding information for a functional product (RNA or a polypeptide)
D. a protein that determines one or more trait
E. a trait that affects phenotype
B and C
C. a section of DNA that contains the regulatory sequences and coding information for a functional product (RNA or a polypeptide)
What is the process of using information in DNA to synthesize RNA?
Transcription
The process of using information in mRNA to synthesize a protein is
Translation
Which of the following is an exception to the “central dogma” of molecular biology?
DNA is the repository of genetic information in all cells.
Messenger RNA is a relatively short-lived "information carrier".
Many genes code for RNAs that function directly in the cell.
Proteins are responsible for most aspects of the phenotype.
All of the above are consistent with the central
dogma.
Many genes code for RNAs that function directly in the cell. Genes code for RNA that function directly, not all RNA codes for proteins.
Which of the following best states the fundamental difference between RNA and DNA?
DNA contains uracil instead of thymine.
Ribose forms a phosphodiester bond in RNA through its 3' carbon; deoxyribose in DNA forms a bond through its 3' carbon.
Ribose has 3 phosphate groups; deoxyribose has none.
RNA contains ribose; DNA contains deoxyribose.
RNA contains ribose; DNA contains deoxyribose. They differ by an oxygen.
What mRNA nucleotide sequence would be transcribed from the DNA sequence 5’ AGCATAGTATC 3’
5'-UCGUAUCAUAG-3'
5'-GAUACUAUGCU-3'
5'-TCGTATCATAG-3'
5'-GATACTATGCT-3'
None of the above
5'-GAUACUAUGCU-3'—B
Which of the following terms accurately describes the genetic code?
A. Ambiguous
B. Overlapping
C. Redundant
A and B
B and C
A and C
All of the above
None of the above
C. Redundant
How many amino acids could 4 nucleotides encode if a codon was 3 nucleotides long?
4
8
12
16
20
64
256
64, 4×4×4
A Frameshift mutation would result from…
deletion of three consecutive bases
a base substitution
a base insertion only
a base deletion only
either an insertion or a deletion of a base
either an insertion or a deletion of a base
The original DNA base sequence is 5’ AGCGTTACCGT 3’
A mutation in the DNA strand results in the base sequence 5’ AGGCGTTACCGT 3’
It may result in a single amino acid change in the protein being coded for by this base sequence.
66)
It is a frameshift mutation.
It is a silent mutation.
It is a deleterious mutation.
It is a frameshift mutation.Extra G in there.
Mutations always decrease the fitness of an organism. True or false?
False.
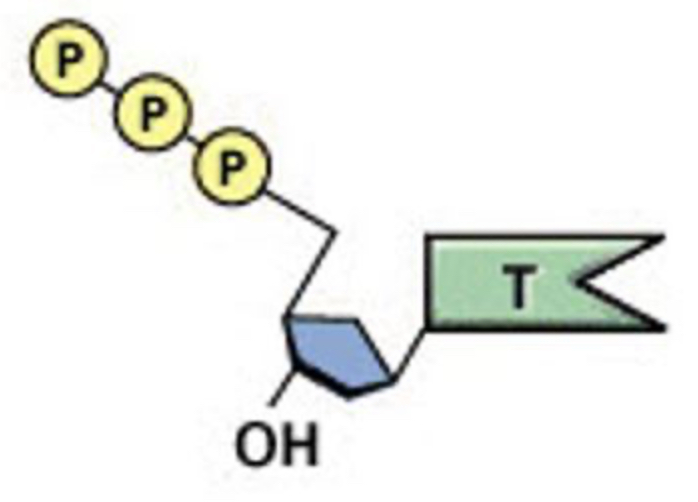
How would this molecule be altered to be used in RNA synthesis?
A. Add another OH to the sugar
B. Remove a CH3 group from the thymine base
C. Remove two phosphates
A & B
A& C
A, B, & C
A and B. You need a RIBOSE sugar, not deoxyribose. So extra OH needed on 2’. It has thymine, no thymine in RNA. We need to change that. When we synthesis we NEED triphosphates, 1 gets incorporated in strand. For synthesis to happen we NEED two phosphate groups to cut off to provide energy that drives synthesis reaction.
A promoter is..
a sequence in DNA that brings RNA polymerase near the site for transcription
a protein that associates with bacterial RNA polymerase to help it bind to DNA
one or more eukaryotic proteins that bind to DNA
near the start of a gene
a sequence in RNA that promotes the release of RNA
polymerase from DNA
a sequence in DNA that brings RNA polymerase near the site for transcription
Which of the following statements best describes the promoter of a protein coding gene?
The promoter is a protein that recruits RNA polymerase.
The promoter is part of the RNA molecule itself.
The promoter is a nontranscribed region of a gene.
The promoter is a site found on RNA polymerase.
The promoter contains the AUG start codon.
The promoter is a nontranscribed region of a gene. Promoter is DNA, anything with protein in answer is wrong. RNA polymerase is a protein. AUG start codon plays a big role in mRNA in TRANSLATION. We aren’t translating here. Promoter is absolutely essential for transcription but is never going to be transcribed :(
A mutation that changes the sequence of nucleotides in a promoter would result in a change in…
the amino acid sequence of the corresponding protein
the base sequence of the corresponding mRNA
the fidelity of translation of the corresponding mRNA
how frequently the corresponding gene is transcribed
how frequently the corresponding gene is transcribed. What is it that a promoter does? It BRINGS RNA polymerase to DNA, changing promoter changes how frequently we transcribe
If you add all the components necessary for transcription to a test tube, which nucleic acids would you find in the test tube after transcription termination?
A. Double-stranded DNA
B. Single-stranded DNA
C. Double-stranded RNA
D. Single-stranded RNA
E. Double-stranded RNA/DNA hybrid
A and D
A and E
B and E
A and D
Which of the following are produced in transcription?
mRNA
IRNA
tRNA
All of the above
None of the above
All of the above
The spliceosome is composed of…
A. DNA
B. RNA
C. Protein
A & B
B& C
A&C
A, B, & C
B and C
SnRNPs…
remove introns from RNA
remove exons from RNA
remove exons from DNA
remove introns from DNA
remove introns from RNA
The DNA shown below is a transcribed part of a gene. The region highlighted in yellow is an intron. What amino acids will be produced?
5’—ATTCGATCCGGCTGACAT-3’
Template:3'—TAAGCTAGGCCGACTGTA-5
Where does translation begin in prokaryotes?
Cytoplasm

Where is the 5’ end of the mRNA in this picture?
ALL nucleic acids are synthesized 5 to 3. We can see the RNA is growing AWAY from the DNA strand. New stuff added to 3, so that means 3 is bottom and 5 is top!
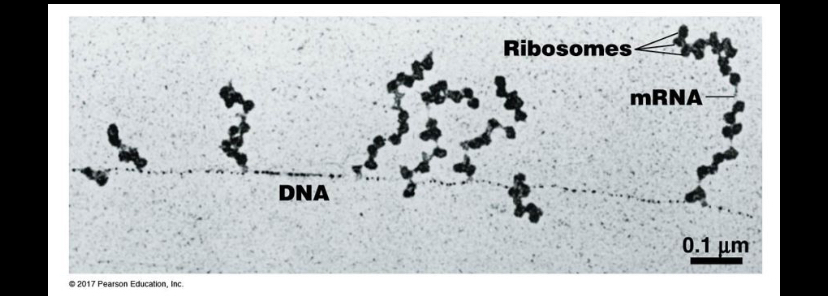
Which direction will the ribosomes move relative to the mRNA?
Downward, in the direction of the DNA strand
Upward, away from the DNA
DOWNWARD. The ribosomes synthesize protein. By moving along RNA 5 TO 3 PRIME, so they can start at 5 and end move to 3. 3 end is attached to DNA, that’s where new nucleotides are being added.
Where does translation begin in eukaryotes?
Cytoplasm
Endoplasmic reticulum lumen
Golgi
Nucleus
Plasma membrane
Cytoplasm. MRNA is taken OUT of the nuclear pore complex and starts translating in cytosol in free ribosome whether we are in pro or eukaryotes.
What kind of molecule serves as an “adapter” that couples amino acid sequences of bases in the mRNA?
DNA
RNA
Protein
Ribosome
RNA, specifically tRNA. 5’ end winds around, 3’ end is where we attach amino acid. Bae pairs between regions of tRNA helps to fold into final structure. Anticodon is part of tRNA. Complementary and antiparallel to codon
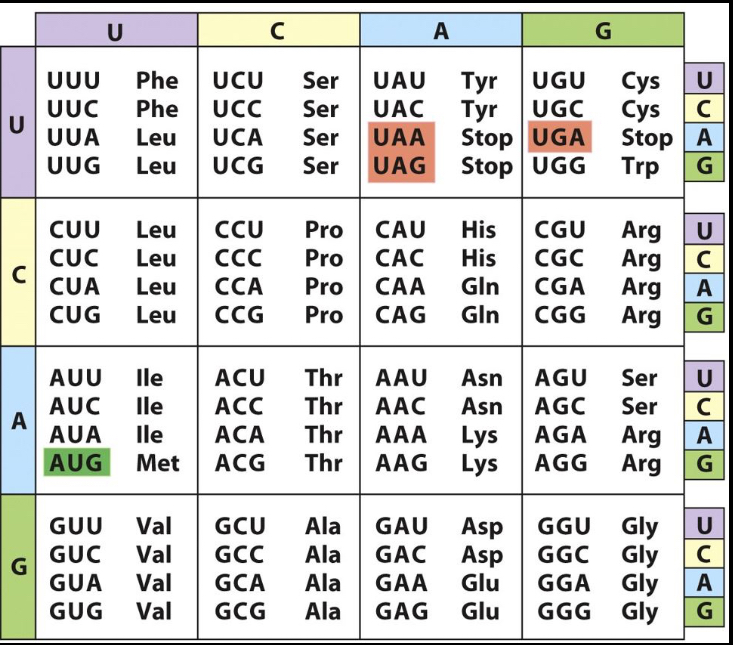
What anticodon would you find on a tRNA carrying Asp?
A. 5'-GAU-3'
B. 5'-GAC-3'
C. 5'-UAG-3'
D. 5'-CUA-3'
E. 5'-AUC-3'
A and B
Original is GAU, and it’s 5 to 3, so tRNA would be antiparallel 3 to 5. But all the options are 5 to 3, so do it backwards! (If og is 3 to 5 no need to flip)
Answer is E
There are sixty one mRNA codons that specify an amino acid, but only 45 tRNAs. This is best explained by the fact that:
competitive exclusion forces some tRNAs to be destroyed by nucleases
the rules for base pairing between the third base of a codon and tRNA are flexible.
many codons are never used, so the tRNAs that recognize them are dispensable
the DNA codes for all sixty-one tRNAs, but some are then destroyed
some tRNAs have anticodons that recognize four or more different codons
C, the rules for base pairing between the third base of a codon and tRNA are flexible. The rules for base pairing at the 3rd position is not always enforced. First 2 have to be perfect, but 3rd has a wobble.
Accuracy in the translation of mRNA into primary structure of a polypeptide depends on the specificity in…
A. binding of the anticodon to the codon
B. binding of the anticodon to the small subunit of the ribosome
C. attachment of amino acids to tRNAs by aminoacy! tRNA synthetases
D. interaction between the aminoacyl-tRNA and the A site of the ribosome
E. A and C
The tRNA needs the right amino acid on it. If we put the wrong one, no further check. We need the right amino acid on the right tRNA by using amino acyl tRNA synthetase.
Anticodon on bottom interacts with the synthetase. There’s specific nucleotides inside that only interact with right synthetase. It attaches the amino acid to tRNA in a very specific way.
What is recognized by an aminoacyl tRNA synthetase?
the nucleotides needed to synthesize a tRNA
one specific codon
the set of redundant codons that specify one amino acid
one amino acid and the set of tRNAs that are coupled to that amino acid
one amino acid and the set of tRNAs that are coupled to that amino acid. In what process do we synthesize tRNA? TRANSCRIPTION. SO A IS WRONG. Codons are in MRNA, so C is wrong. So D is the right answer.
Ribosomes consist of…
A. DNA
B. RNA
C. Protein
D. DNA & RNA
E. RNA & protein
F. DNA, RNA, and protein
E. RNA and protein. RIBOZYMES catalyze the peptide bond between amino acids in ribosomes. RNA component of the ribosome is what helps carry out catalysis.
ALSO. AT ANY given moment, 3 tRNAs can be working and interacting with the ribosome.
How does bacterial ribosome recognize where to start translation?
The small ribosomal subunit binds to the 5' cap of the mRNA.
The small ribosomal subunit binds to the start codon.
The small ribosomal subunit binds to the 3' poly(A) tail of the mRNA.
The small ribosomal subunit binds to a sequence in the mRNA just upstream of the start codon.
The small ribosomal subunit binds to a sequence in the mRNA just upstream of the start codon. Once tRNA is in place, large subunit joins.
In the context of protein synthesis, what is meant by translocation?
The completed polypeptide is released from the ribosome.
The ribosome slides one codon down the mRNA.
The two ribosomal subunits are joined in a complex.
The polypeptide chain grows by one amino acid.
The ribosome slides one codon down the mRNA. Ribosomes slides along mRNA, movement of one relative to the other.
What recognizes a stop codon in the A site?
A stop tRNA with a stop anticodon
The small subunit of the ribosome
A protein called release factor
Nothing. The presence of the stop codon causes the ribosome to dissociate from the mRNA.
A protein called release factor.
Is there a stop amino acid? NOPE. Is there a tRNA that has an anticodon for a stop codon? NOPE. STOP codons are NOT recognized by tRNAs, but by proteins.
Release factor will specifically recognize the stop codon.
What happens to mRNA after the termination of translation in eukaryotes?
The mRNA is spliced, and a 5' cap and polyA tail are added.
The mRNA remains in the cytosol and may be translated again.
The mRNA moves through the nuclear pore out of the nucleus.
The mRNA remains anchored on the membrane of the rough ER.
The mRNA is immediately degraded, and its components are recycled and used in the synthesis of new RNA molecules.
The mRNA remains in the cytosol and may be translated again.
Does ANYTHING change about the mRNA in translation? NO. MRNA is the same as it went in. Ribosome comes out same as it went in. TRNAs have transferred amino acid.
MRNA will remain in cytosol so process can keep going. We can make a ton of protein from just 1 mRNA molecule.
Which of the following are examples of post-translation modifications of proteins?
A. addition of a lipid group
B. glycosylation
C. phosphorylation
A and B
A and C
B and C
All of the above
All of the above. We know kinases add phosphate group. We can add a lipid group to anchor proteins at membranes. We can also add sugar groups like mannose 6 phosphate to go to lysosome! These are all examples of modifications.
Why do DNA and RNA move toward the positive electrode in electrophoresis?
The phosphate groups in DNA and RNA are positively charged.
The phosphate groups in DNA and RNA are negatively charged.
The nucleotide bases in DNA and RNA are positively charged.
The nucleotide bases in DNA and RNA are negatively charged.
The phosphate groups in DNA and RNA are negatively charged.
We load samples, turn on charge, our samples will run to positive charge. Negative on top, + on bottom.

The DNA fragment indicated is approximately ___ base pairs in size.
300
350
580
600
700
Numbers on left show base pair amount! 580!
Protein samples are usually treated with charged detergent before they are run on a gel. Under these conditions, protein gel electrophoresis separates proteins on the basis of…
charge of the side chains
fluorescence
size
solubility
tertiary structure
SIZE. SDS coats everything with negative charge. Proteins have broken disulfide bonds, boiled, coated with negative charge and then we can separate on gel.
Which of the following would move the farthest on a protein gel?
cyclin dependent kinase (34 kDa)
insulin receptor (156 kDa)
phosphofructokinase (85 kDa)
sodium-potassium pump (100 kDa)
CDK cuz it’s small. Small things move farther on gels.
During PCR, the purpose of raising the temperature of the solution to near boiling temperatures is to…
increase the rate of reactions for DNA polymerase
decrease the rate of reactions for DNA polymerase
separate the two parental DNA strands
break the DNA into short segments with exposed sticky ends
separate the two parental DNA strands
After 30 rounds of PCR, approximately how many copies of the target region will be made, assuming 100% efficiency?
2 ^ 30, about a billion.

You use the DNA sequence and primer below for a PCR reaction. What primer would you need to amplify the other strand?
5'-CCCTGGGCT-3'