M2L4 Endomembranes and vesicular transport
1/28
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
29 Terms
Where are polarised vs non-polarised cells located?

Polarised - usually when sitting against an interface (eg. air-water interface in the lung)
Non-polarised - cells within an organ
What factors affect the properties of the lipid bilayers of membrane-bound organelles in a secretory pathway?
Membrane bound organelles are encompassed by lipid bilayers which may have variable composition depending on the role of the organelle in the secretory pathway
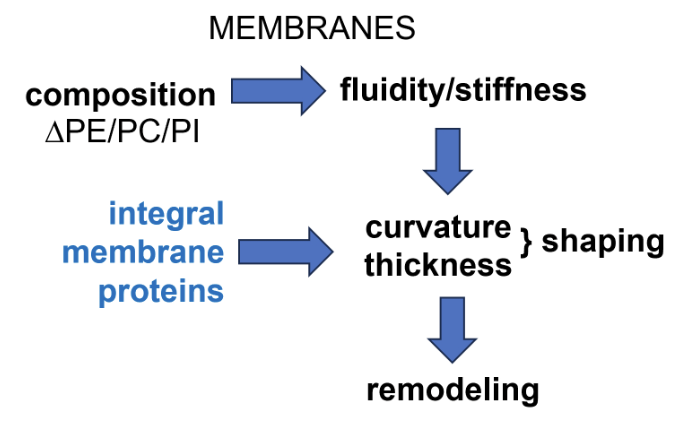
Balance of different phospholipids (eg. phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol) controls membrane fluidity/stiffness
Integral membrane proteins controls the curvature and thickness of membranes, thereby regulating its shaping
This altogether controls how the membrane can be remodelled
Explain the multi-directional flow of cargo via the secretory pathway
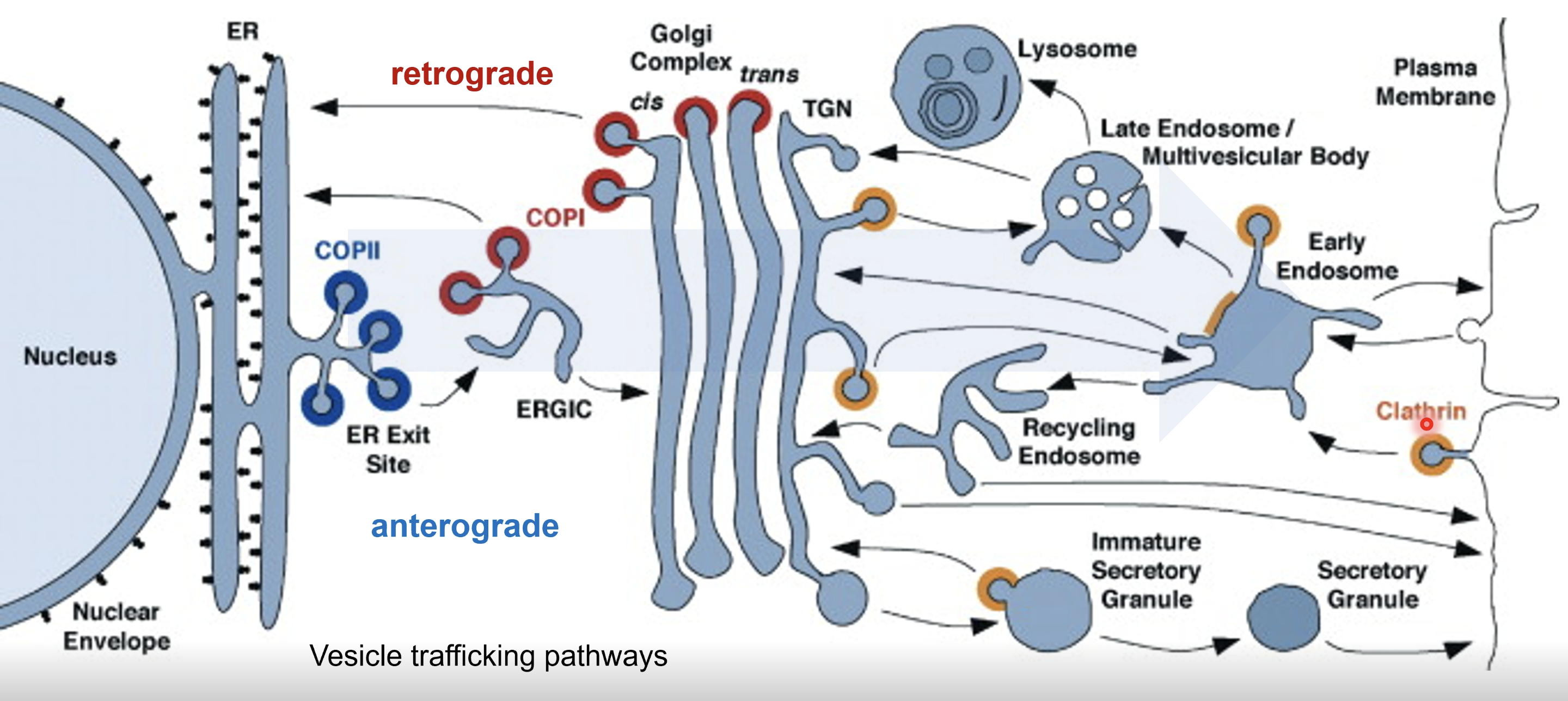
rER synthesises proteins that are to be exported from the cell
COPII vesicles mediate forward traffic, therefore proteins synthesised in the ER are packed into COPII vesicles
COPII vesicles fuse toegther to form the ERGIC transition cpmpartment before fusing with the cis Golgi
Cargo moves from the cis through to the trans Golgi, undergoing post-translational modifications and sorting
From the trans Golgi network (TGN) the cargo is sent to the plasma membrane for secretion, endosomes which may direct cargo to lysosomes or to the plasma membrane, or secretory granules
COPI vesicles mediates backward traffic from the Golgi to ERGIC to the ER to retrieve mis-sorted ER resident proteins and Golgi enzymes that need to remain in earlier compartments
Clathrin coated vesicles are involved in transport from TGN to endosomes (delivery to lysosomes or recycling) or plasma membrane to endosomes (endocytosis)
Early endosome - first stop for incoming vesicles from the plasma membrane
Recycling endosome - returns useful materials to plasma membrane
Late endosome/multivesicular body - delivers cargo to lysosomes for degradation
How is the ER shaped?
ER consists of sheets/cisternae (flatter regions studded with ribosomes), tubules (highly curved tubular membranes), and 3-way junctions (where tubules intersect and form a network)
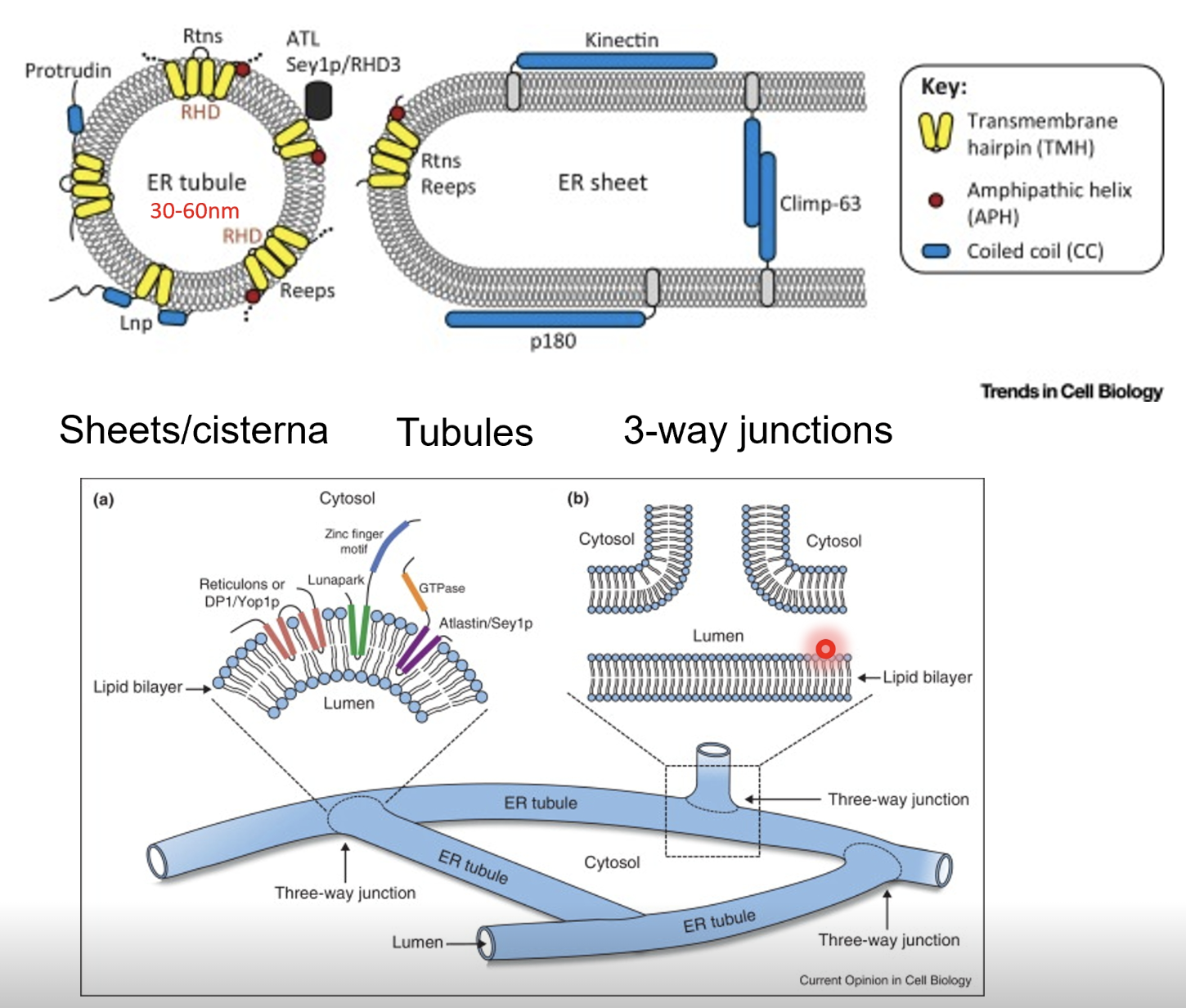
What factors affect the shape of the ER?
Shape of the ER (sheets vs tubules) is determined by membrane curvature and embedded proteins
Reticulons (Rtns) and receptor expression enhancing proteins (REEPs) insert into the membrane as wedge-shaped dimers or hairpin loops
These proteins embed into the outer leaflet of the lipid bilayer, causing local bending and tubule curvature
Curvature stabilising proteins (eg. Climp-63, p180. kinectin etc) act as luminal spacers or cross0bridges across the ER membrane to maintain sheet spaciong, preventing collapse or fusion of membranes to stabilise flat cisternal regions
Amphipathic helices insert shallowly into the cytoplasmic leaflet of the bilayer, adding asymmetry to promote membrane bending, thus fine tuning the curvature
The ER network is highly dynamic - tubules, extend, retract and fuse, driven by motor proteins (eg. kinesin) interacting with microtubules
Atlastin is a GTPase that mediates fusion at 3-way junctions to maintain ER network continuity
How is the ER connected to mitochondria?
The ER and mitochondria are tightly linked by membrane contact sites, where tethering protein complexes form “funnel-like” bridges that allow direct lipid and calcium transfer between them
ER-mitochondrial encounter structure (ERMES) is formed by Mmm1, IP3R, and MCU (ER membrane protein) and Mdm10, Mdm12, and Mdm34 (mitochondrial membrane proteins)
What are the features of the ER lumen?
Oxidative environment
High Ca2+ concentration (x1000 cytosol)
Near neutral pH (~7.2)
What is the importance of the oxidative environment in the ER lumen?
The cytosol is a reducing environment due to molecules like GSH that keep cysteine resides in their reduced (-SH) form, making it hard to form disulfide bonds
The extracellular space is an oxidative environment in contrast, allowing disulfide bonds to form (-SH + -SH —> S-S) which stabilise proteins, especially those secreted or at the cell surface
Therefore the ER which synthesises secretory or membrane proteins will recreate and oxidative environment to allow disulfide bonds to form for proper protein folding/structure
What is the importance of high Ca2+ concentration in the ER lumen?
Calcium releases an important signalling mechanism either from outside the cell or in the ER in muscular contraction
Number of proteins are calcium-dependent
What are the features of the ER membrane?
High PE, PC, PI
Low cholesterol (changes the rigidity of the membrane), though cholesterol is manufactured in the ER
Makes ceramides
How are lipid droplets formed by the ER?
Lipid droplets (LD) are formed from the ER lipid bilayer by the concentration of neutral lipids, eg. diacyl- and triacylglycol, and cholesterol esters
Proteins in the ER membrane bring diacyl- and triacylglycerol in proximity and concentrate them in the ER membrane to form a lens (local bulge of lipids) which buds off
As LDs are formed in the centre of the lipid bilayer, when they bud off they only have a single membrane (heads face outwards and tails face inwards, holding/stabilising the LD)
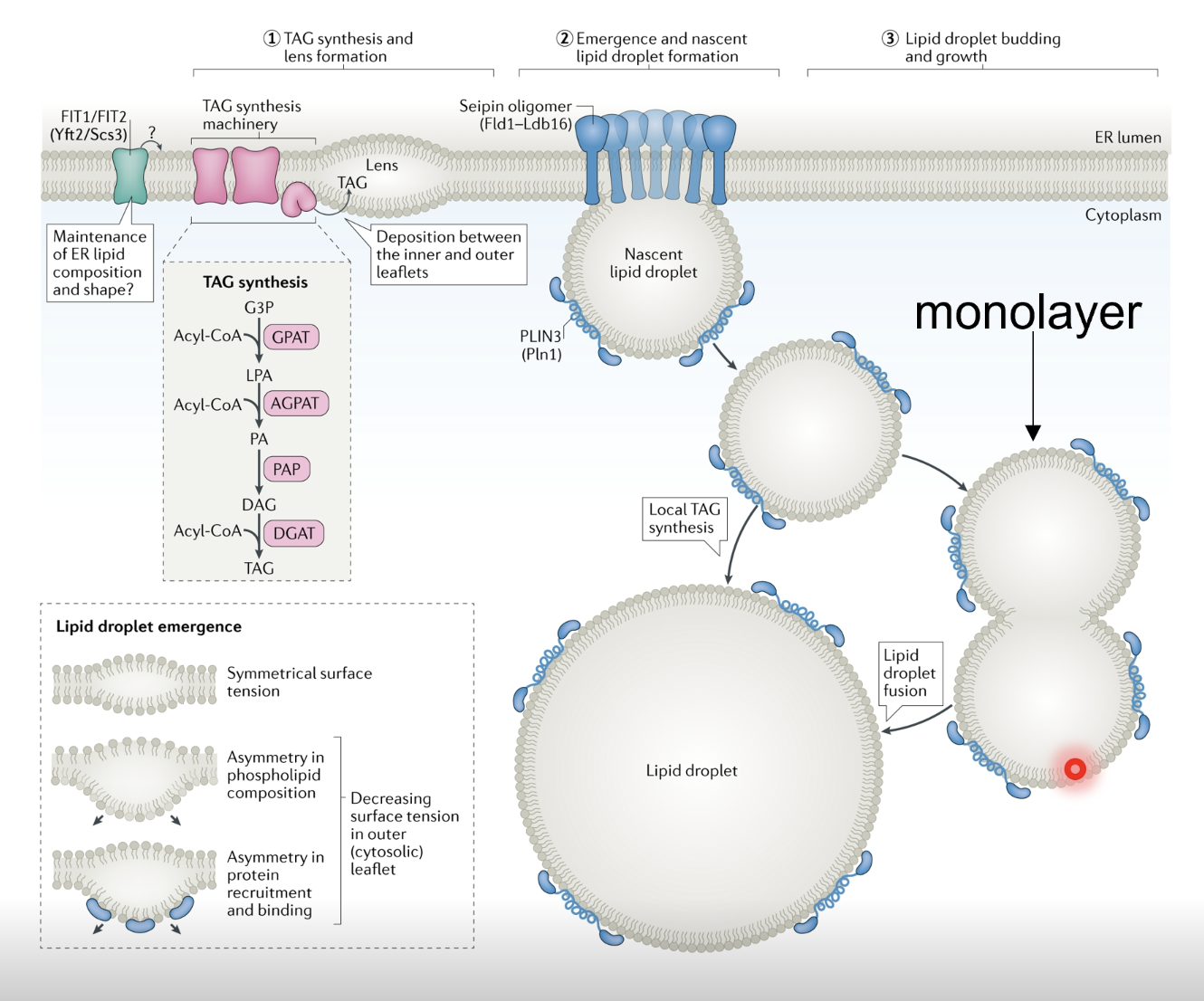
What is the importance of LDs?
Important storage mechanism of lipids
If a cell needs lipids during starvation it can release it from LDs
LDs can make contacts with organelles and are important in lipid trafficking and energy homeostasis
Explain the biogenesis of secreted and integral membrane protein cargo at the ER
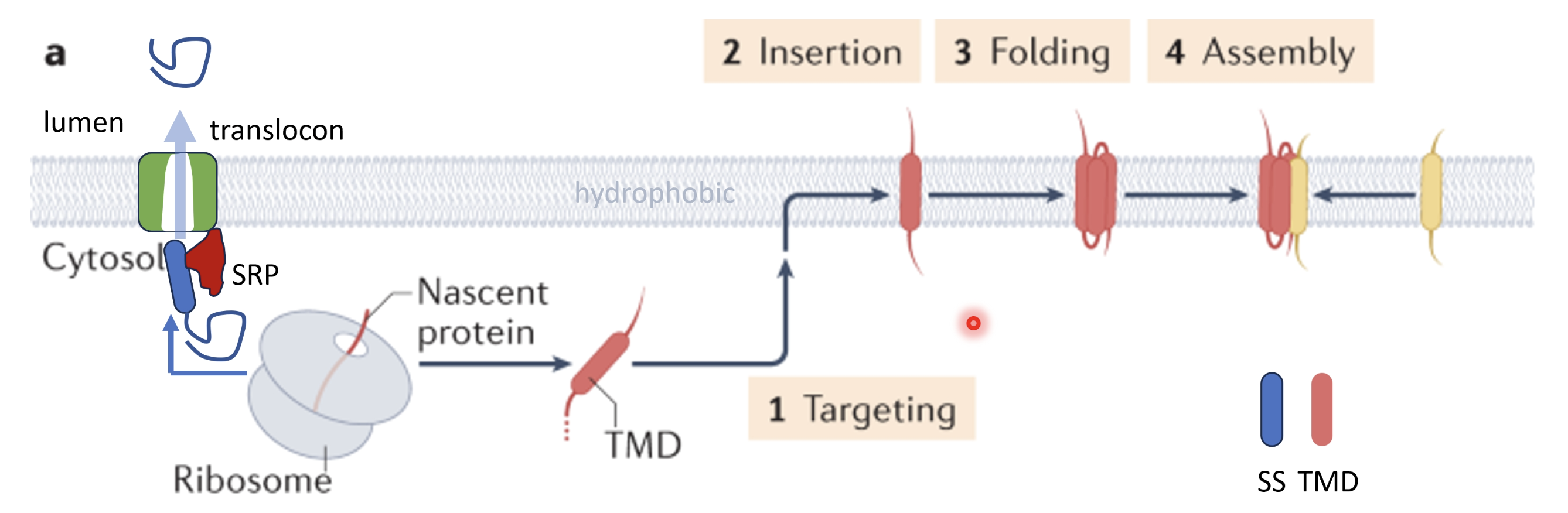
Translation begins in the cytoplasm through free ribosomes
The mRNA encodes a (~18-25 aa long) hydrophobic N-terminal ER signal sequence which is recognised by the signal recognition particle (SRP) once it emerges in the nascent polypeptide which pauses translation temporarily and targets the complex to the ER membrane, docking to the SRP receptor and Sec61 translocon
Upon docking of the ribosome to the ER translation resumes and SRP is released, threading the growing polypeptide directly into the Sec61 channel either completely through into the ER lumen (for secreted proteins) or partly inserted into the membrane for membrane proteins
For proteins destined for the ER lumen, the signal sequence is cleaves after the polypeptide is threaded into the lumen, releasing it into the ER
For a transmembrane protein, there will be an N-terminal start transfer sequence and hydrophobic transmembrane domains (21-25 aa) which cause the polypeptide to be released laterally, remaining anchored to the lipid bilayer
What types of membrane bound proteins can be synthesised at the ER?
Integral proteins
Monotopic
Type I (single-pass, N-lumen/C-cytosol) - N terminal signal sequence is cleaved off
Type II (single-pass, N-cytosol/C-lumen) - First hydrophobic region acts as both signal anchor and membrane anchor (not cleaved), and positive residues dictate orientation
Polytopic - multiple TMDs alternate across bilayer, each hydrophobic stretch acts as a start- or stop-transfer sequence
Tail-anchored (TA) proteins - single C terminal transmembrane domain, hydrophobic anchor emerges after translation
GPI-anchored proteins - protein that is attached to the cell membrane by a glycosylphosphatidylinositol (GPI) glycolipid anchor
positive inside rule
Orientation of integral proteins follow the ‘positive inside’ rule - regions with positively charge residues (lysine (K+) and arginine (R+)) stay on the cytosolic side
Applies most clearly and predictably to Type II transmembrane proteins but it also influences the orientation of multi-pass and other single-pass membrane proteins
Explain the steps how hydrophobic sequences are a key driver of membrane protein topology
At a certain frequency hydrophobic sequences do not like to be in an aqueous environment and may thus spontaneously insert into the lipid bilayer
Often the insertion of the hydrophobic sequences into the bilayer needs to be facilitated
Channel (Sec61) mediated - ball and chain gated channel, SRP binding displaces the ball and opens the lateral gate, allowing the hydrophobic sequence to be threaded through
For TA proteins, SRP falls off and can not carry them to the ER, instead its localisation is mediated by the GET and EMC complexes
These complexes have components with hydrophobic grooves in soluble protein, which in turn have cofactors that are ER-bound
When hydrophobic sequence of the TA protein emerges so late it gets bound by hydrophobic grooves which then coordinate with the membrane insertion mechanism, delivering it to the secondary insertion pathway
For signal anchor proteins (eg. GPCRs), they have difficulty getting into the ER as their N-terminus resembles a type II protein
They use EMC to set their first TM domain before getting transferred to Sec61
What is a reason for energetic instability when synthesising multi pass membrane proteins (eg. β₁-adrenergic receptor)?
When folding multi pass membrane proteins (eg. β₁-adrenergic receptor) the transmembrane domains contain some polar or charged residues which is energetically unstable
Intramembrane chaperone complexes shield the hydrophilic residues while other transmembrane domains are being synthesised
Once all the transmembrane domains are synthesised the hydrophilic residues across these domains can form interactions to stabilise each other
PAT, BOS, and GEL chaperone complexes coordinate with Sec61 to provide stability
How do molecular chaperones help in protein maturation in the ER?
Molecular chaperones (particularly HSP70 and HSP90) help to either actively or passively aid in protein folding within the ER (and also in the cytosol)
Passively - binding hydrophobic domains that would be buried
Actively - by remodelling
Chaperones are mostly soluble proteins which are assisted by J domain proteins which activate BiPs (an HSP70 protein), the main factor for protein folding in the ER
BiP uses its ATPase activity to carry out its function
Calnexin (membrane-tethered, binds oligosaccharides) and calreticulin (soluble) are ER chaperones which assist in the folding of glycoproteins alongside BiP while
Also serves an interrogration function to assess whether a protein is folded by grabbing on to its glycan and there are domains which sense folding status
Follows a cycle of addition, modification and release of these terminal glucoses to interact, disengage, and re-engage as a quality control measure for glycoproteins
If they pass QC they get released, if not they are degraded
What is the role of protein disulphide isomerases (PDIs) in protein maturation in the ER?
Protein disulphide isomerases (PDIs) catalyse formation of disulphide bonds between sulfhydryl groups of cysteine residues to give an extra level of structure and aid in folding
What is the role of peptidyl prolyl isomerases in protein maturation in the ER?
Peptidyl prolyl isomerases catalyse the cis-trans isomerisation of peptide bonds preceding proline residues to aid in protein folding
What PTMs take place in the ER?
Signal peptide cleavage by signal peptidase complex
Asparagine (N)-linked oligosaccharides - through the canonical sequence of N-X-S/T, regulated by oligosaccharyl transferase complex which adds large sugar groups to the proteins (important for recognition, binding, folding)
Disulphide bond formation
Trimming of sugar chains after glycosylation by glycosidases as part of ER quality control and folding cycles
What are two modes of ER export?
Bulk flow involves non-specific selection of cargo when forming budding vesicles
Alternatively specific cargo receptors can bind motifs that bring proteins into emerging vesicles
How do vesicles accommodate variably sized cargo?
Sec31 forms cages which shape the vesicles
Cages are adaptable, able to stretch to accommodate variably sized cargo which is dictated by different receptors (eg. TANGO1)
Explain vesicle trafficking and directionality mediated by Rabs and SNAREs
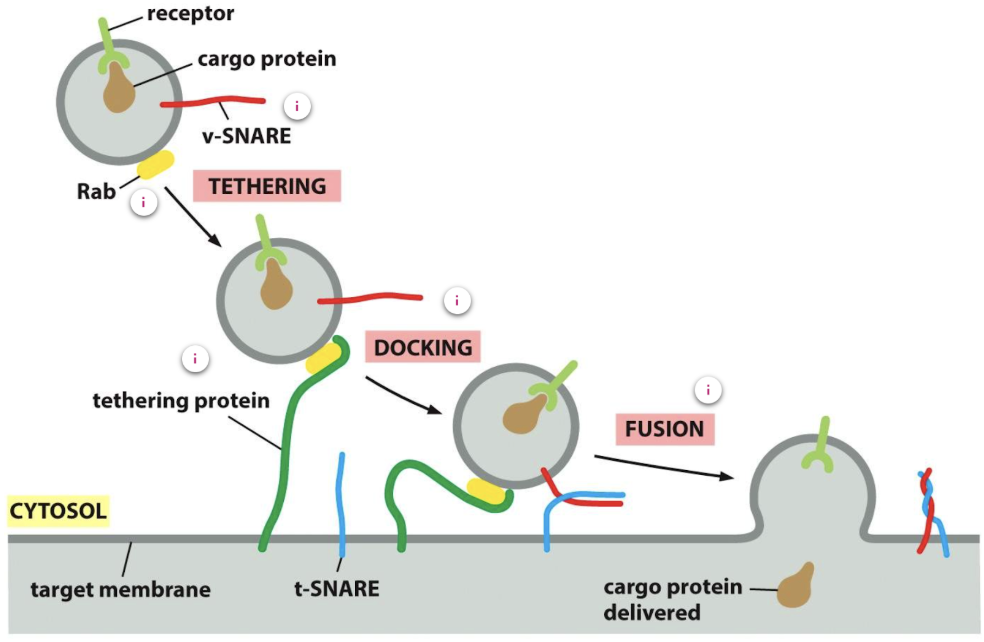
Trafficking and directionality is mediated by Rabs and SNAREs
Rab proteins ensure that the vesicle fuses with the correct membrane as each organelle and vesicle has a distinct combination of Rab proteins on its membrane
At the correct membrane Rab-GEF exchanges GDP for GTp to activate Rab
Rab-GTP recruits Rab effectors - filamentous tethering protein of the target membrane which binds to the Rab protein to allow the vesicle to dock
Once a tethering protein captures a vesicle by binding to a Rab protein on its surface, v-SNAREs (on the vesicle membrane) interact with complementary t-SNARES (on the target membrane) to allow firm docking, catalysing membrane fusion
Explain ER-associated degradation (ERAD)
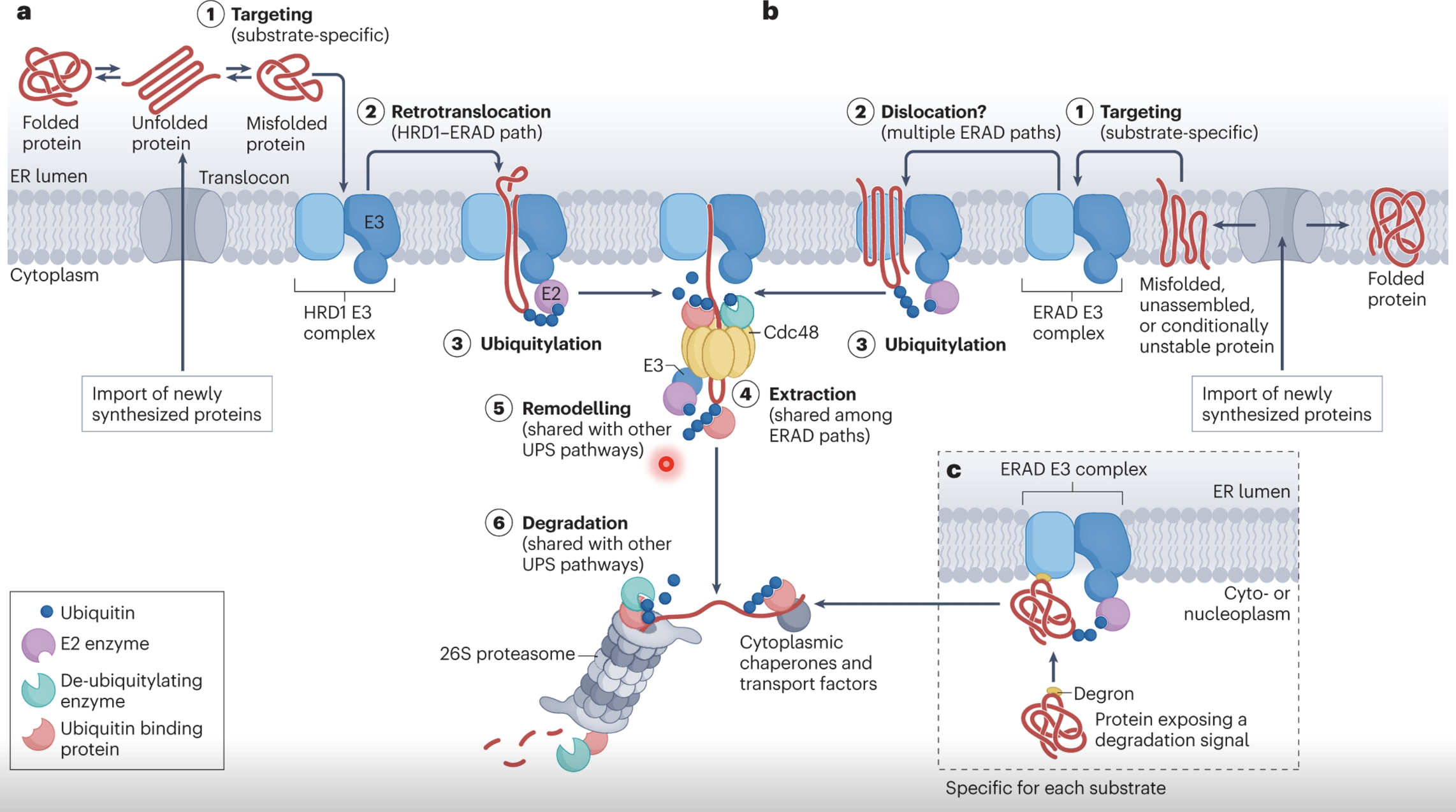
Misfolded proteins have propensity to aggregate and cause cell toxicity, therefore they must be degraded via ER-associated degradation (ERAD)
Misfolded proteins expose degrons - hidden hydrophobic patches, unpaired cysteines, or abnormal glycans which signal for degradation
Specialised recognition factors bind to defective proteins and direct them to and ERAD E3 ubiquitin ligase complex (eg. HRD1)
Once targeted the misfolded protein must be extracted from the ER lumen/membrane into the cytosol via retrotranslocation, mediated by HRD1 and other accessory proteins
As the protein is extracted it is also polyubiquitinated
Cdc48 uses ATP hydrolysis to physically extract the misfolded protein and release it in the cytosol
Deubiquitinating enzymes (DUBs) may trim or remodel ubiquitin chains to prevent premature degradation, guiding the substrate to the proteasome
The protein is recognised by the 26S proteasome which unfolds and threads the protein through its catalytic core for degradation/recycling
What is another mechanism for degrading misfolded proteins apart from ERAD?
Apart from ERAD, misfolded proteins could also be sent to distal sites via ER exit sites, though this may cause more damage due to the protein being defective or harmful
ERLAD (direct ER to lysosome associated degradation) and ER-phagy could also be used to target protein aggregates (degrading altogether rather than having to thread through the proteasome)
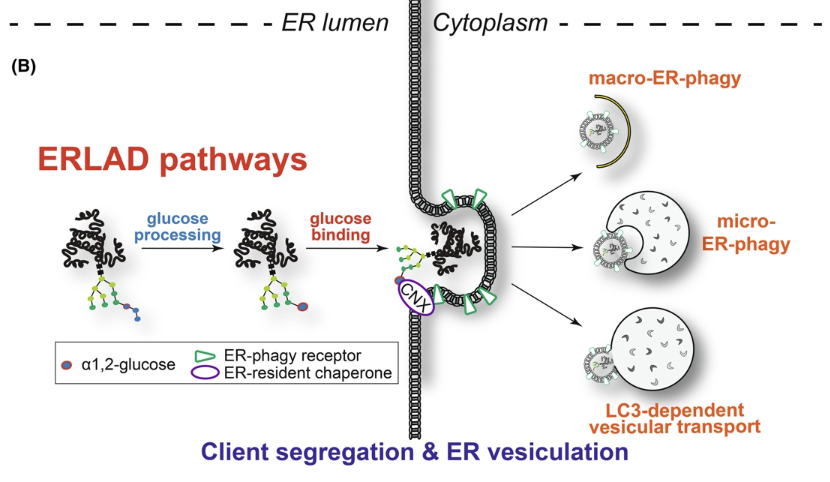
How is proteotoxic stress generated in the ER?
If the capacity for protein folding is outstripped by the amount of unfolded/misfolded proteins it can cause proteotoxic stress
Particularly in the ER especially due to proteins that require BiP
Explain the proteotoxic stress response in the ER
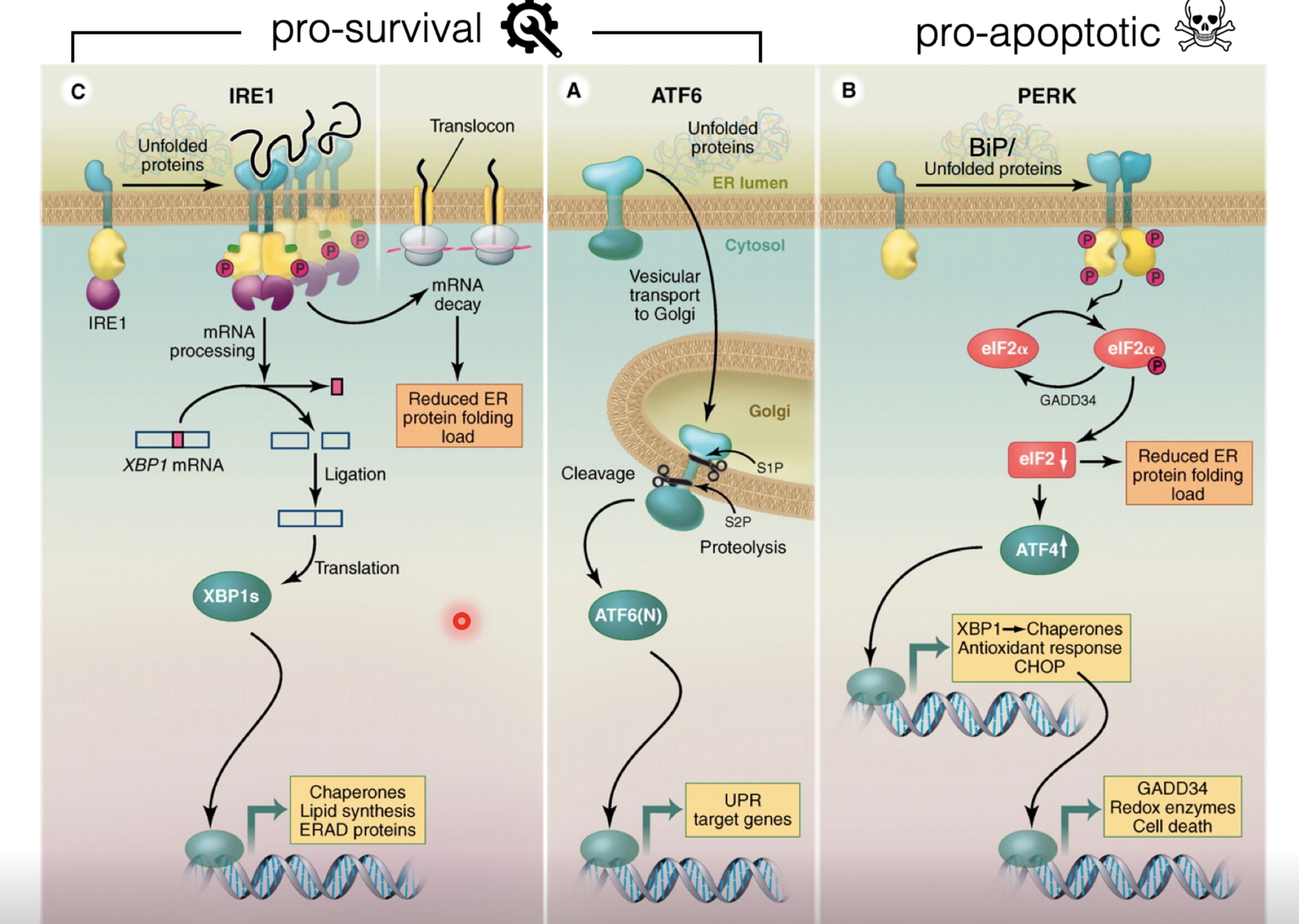
Un/misfolded proteins in the ER compete with unfolded protein transducers IRE1, ATF6 and PERK (act as sensors which bind BiP)
When bound to BiP they can not sense stress (inactive)
When there is more misfolded protein than soluble BiP, BiP is displaced to bind the misfolded proteins, thus activating the transducers
Initial response is pro-survival
Activated IRE1 has endonuclease activity which splices out a region of an mRNA to produce a transcription factor XBP1 which upregulates genes for chaperones, lipid synthesis, and ERAD proteins
Active ATF6 is released to the Golgi and gets cleaved by cyclin proteases, releasing the transcription factor to upregulate unfolded protein response (UPR) target genes
Folding machinery and degradation machinery is upregulated to resolve the stress and ER size is also increased to dilute the misfolded proteins
If the damage is too severe or in cases of chronic stress, activated PERK triggers apoptosis
PERK monomers dimerise which unlocks its kinase activity, allowing it to phosphorylate eIF2α, attenuating global translation (to reduce the load of new proteins entering the ER), but allowing selective translation of ATF4
ATF4 induces genes encoding XBP1 and chaperones as well as antioxidant response genes to mitigate stress, however CHOP is also induced to cause apoptosis with persisting stress
How does ER stress relate to cancer?
Constitutive upregulation/activation of ER stress response pathways to tolerate higher levels of misfolded protein, shifting the balance to favour pro-survival
UPR is a model of non-oncogenic addiction, it is not a driver of proliferation or cancer but is a mechanism to handle stress and manage a non-optimal environment
Sometimes can cause therapeutic resistance
Some evidence suggests that the QC pathways in the ER could be targeted for therapy - eg. KD of IRE1-XBP1 branch of the UPR decreased tumour burden significantly in mice