Eukaryotic Transcription
1/31
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
32 Terms
Steps of Eukaryotic Transcription
Initiation- Transcription factors bind to the promoter and recruits RNA polymerase ll. Then DNA unwind, and transcription begins at the +1 site.
Elongation- RNA polymerase II moves along the DNA from 5' to 3' direction making a complementary mRNA strand. Then it adds nucleotides one by one and forms phosphodiester bonds to build the RNA chain.
Termination: Continues RNA polymerase does not stop immediately at the end of the gene. It keeps going 1,000–2,000 base pairs past the termination signal. Then the RNA transcript is cleaved (cut) and the mRNA is released.
Types of Eukaryotic RNA Polymerase
RNA polymerase l
RNA polymerase ll
RNA polymerase lll
RNA polymerase
Found in the nucleolus
Synthesizes rRNA
Makes up the core structure and catalytic components of ribosomes
rRNA
A type of molecule that combines with proteins to form ribosomes, the cellular machinery responsible for making proteins
RNA polymerase ll
Found in the nucleus
Transcribes protein-coding genes (pre-mRNA). Then pre-mRNAs undergo processing before translation.
Responsible for majority of transcription activity
RNA polymerase lll
Found in the nucleus
Transcribes tRNAs and small nuclear RNAs
These molecules play roles in translation and RNA processing
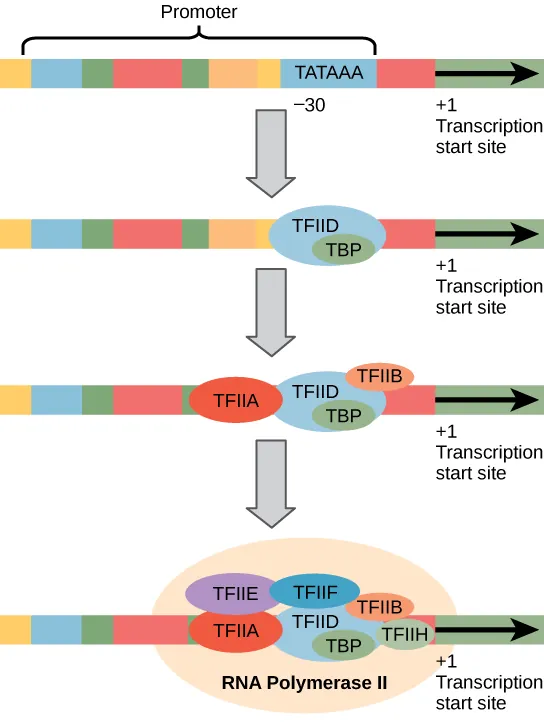
Eukaryotic Promoter
The promoter is a DNA sequence upstream (before) the transcription start site (TSS), which is labeled as +1.
“Upstream” means in the negative direction (−) from the start site.
This region is where RNA polymerase and transcription factors bind to begin transcription.
TATA box (TATAAA): at -25 to -35 bp, A-T rich for easy unwinding
Eukaryotic Transcription Factors (TFs)
Basal transcription factors (TFIIA-TFIIJ) assemble at promoter
ALL need to bind before initiation
RNA polymerase ll then binds and forms transcription initiation complex
Eukaryotic Transcription Factors (TFs) Functions
To bind to DNA
Stabilize pre-initiation complex
Unwind the DNA
Eukaryotic Initiation
Transcription factors (TFs) bind first to the promoter (pre-initiation complex)
TFs help recruit RNA polymerase ll to start transcription (initiation complex)
RNA polymerase cannot bind DNA alone
TATA box: Consensus sequence for eukaryotes
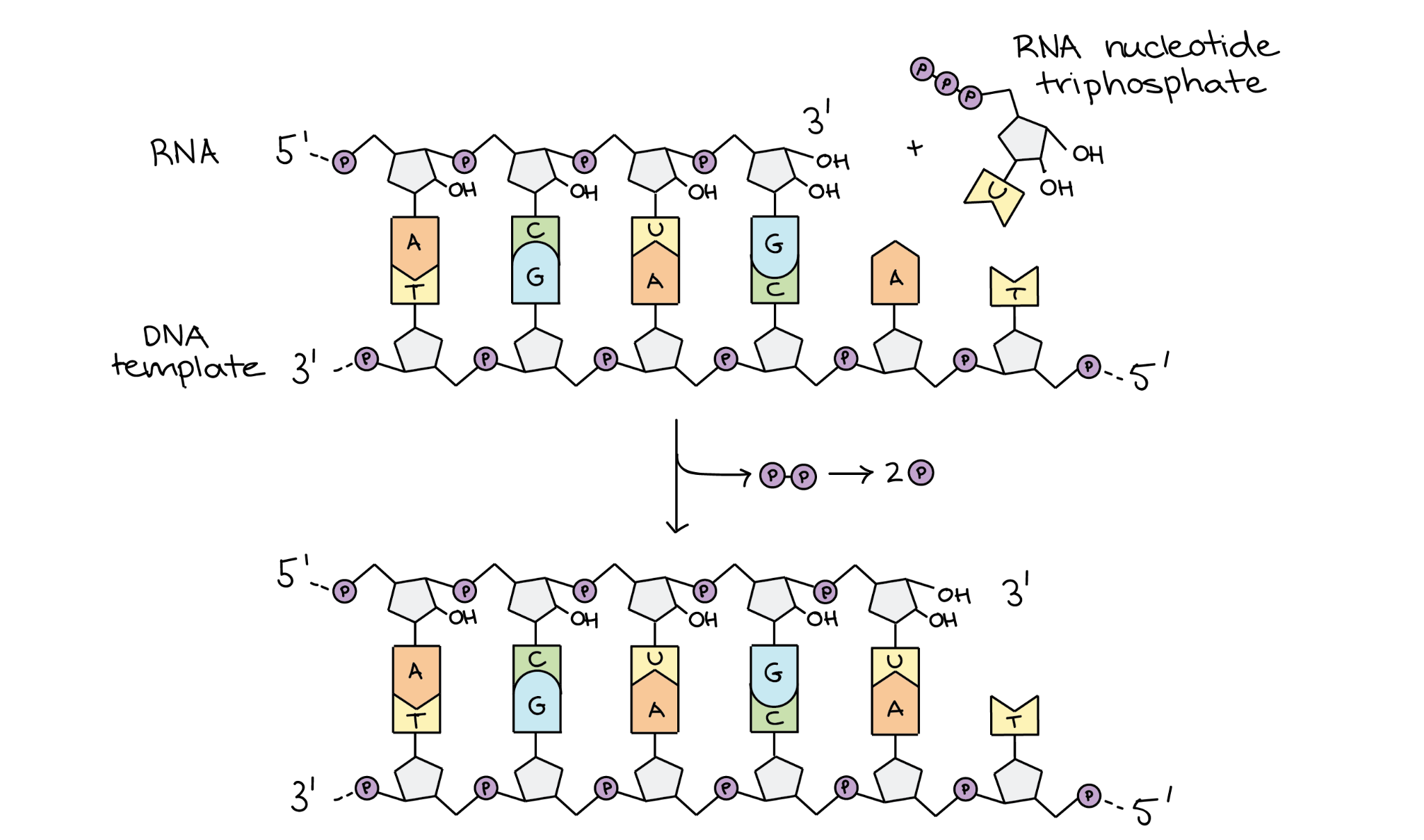
Elongation
Same as prokaryotic elongation
RNA polymerase II moves along the DNA from 5' to 3' direction making a complementary mRNA strand.
It adds nucleotides one by one and forms phosphodiester bonds (links between the nucleotides) to build the RNA chain.
Termination
RNA polymerase does not stop immediately at the end of the gene.
It keeps going 1,000–2,000 base pairs past the termination signal.
Then the RNA transcript is cleaved (cut) and the pre-mRNA is released.
Eukaryotic Pre-Transcriptional Regulation
Chromatin status
Basal and other transcription factors
Enhancers- turn on transcription and this is where activators bind
Silencers (Repressers)- turn off transcription and this is where repressers bind as well
DNA Structure
DNA Double Helix: Composed of nucleotide base pairs (A-T and G-C)
Chromatin: DNA wrapped around histone proteins → forms nucleosomes
Chromosomes: High organized, and compact structures of chromatin found in the nucleus
The structure of DNA determines accessibility for transcription to occur
Chromatin
DNA wrapped around histone proteins → forms nucleosomes
Chromosomes
High organized, and compact structures of chromatin found in the nucleus
2 Types of Chromatin status
Heterochromatin: “Closed” Chromatin
Tightly packed
No access to DNA= No transcription
Euchromatin: “Open” Chromatin
Loosely packed
Access to DNA= Transcription
Transcription occurs only in euchromatic regions of chromosomes
Heterochromatin
“Closed” Chromatin
Tightly packed
No access to DNA= No transcription
Euchromatic
“Open” Chromatin
Loosely packed
Access to DNA= Transcription
Transcription occurs only in euchromatic regions of chromosomes
Epigenetic Regulation of Transcription
Histone modifications: Chemical modifications to the histone proteins
Histone Acetylation: Loosens chromatin → activates transcription
Histone Methylation: Tightens chromatin → silences transcription
DNA methylation
Adds methyl groups to the DNA strand
Tightens chromatin → silences transcription
Other Transcription Factors
Transcriptional Activator Proteins
Transcriptional Repressor Proteins
Coactivators and Corepressors
Function of Transcriptional Activator Proteins:
Bind to specific DNA sequence (often enhancers)
Recruit or stabilize the transcription machinery
Increase the rate of transcription initiation
Function of Transcriptional Repressor Proteins
Bind to specific DNA sequences (often silencers/repressors)
Block activator binding or inhibit RNA polymerase activity
Function of Coactivators and Corepressors
Do not bind DNA directly
Act as bridges between enhancers/repressors and the transcription complex
Can modify chromatin structure
Enhancers
DNA sequences that increase transcription efficiency
Can be upstream, downstream, or within the gene
Bind transcriptional activator proteins (TF) → DNA looping brings them to the promoter
Helps stabilize RNA polymerase ll
Can have multiple enhancers to control transcription
Silencers (Repressors)
DNA sequences that can lead to transcription block
Bind transcriptional repressor proteins → DNA looping brings them to the promoter
Prevent activators or TFs from binding
Can recruit histone deacetylases to tighten chromatin
Deacetylases
An enzyme that removes acetyl groups from histone proteins
Eukaryotic pre-mRNA Processing
Eukaryotic genes contain exons(coding) and introns(noncoding)
Introns must be removed and exons joined to create a continuous coding sequence
mRNA processing increases RNA stability and lifespan:
Eukaryotic mRNA: lasts hours
Prokaryotic mRNA: lasts seconds
Pre-mRNA is protected by RNA-stabilizing proteins during
Three Main Steps of mRNA Processing
5’ Capping
3’ Poly-A Tail Addition
Splicing (intron removal)
Together, these modifications produce a stable mature (spliced) mRNA
5’ Cap and 3’ Poly-A Tail
5’ Cap
Added to the 5’ UTR (untranslated region) while transcription is still occuring
Protects mRNA from degradation
Helps ribosomes recognize and bind to the mRNA during translation initiation
3’ Poly-A Tail
Poly-A polymerase adds 200 adenine nucleotides to the 3’ UTR (untranslated region)
Protects mRNA from degradation
Aids in nuclear export and translation
mRNA Splicing
Introns are removed and exons are joined by the spliceosome
Spliceosome: Protein, enzyme, and RNA complex that removes introns and splices exons together
Alternative Splicing
Different combinations of exons are joined together (spliced) to produce multiple mature mRNA transcripts from a single gene