BIOC*2580: Enzymes and Reaction Rates
1/38
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
39 Terms
nucleophilic catalysis
Enzymes can speed up reactions by providing a better nucleophile.
e.g. Cys-SH, His-N: Asp or Glu -COO:-
More rarely Tyr or Ser -:OH or Lys :NH2
electrophilic catalysis
An electrophile is an electron-seeking group; there are no really good electrophilic amino acids.
An enzyme may contain a non-amino acid helper molecule called a prosthetic group as part of its structure (cofactors/coenzymes). They bind at the enzyme catalytic site and initiate reaction by withdrawing electrons from the substrate.
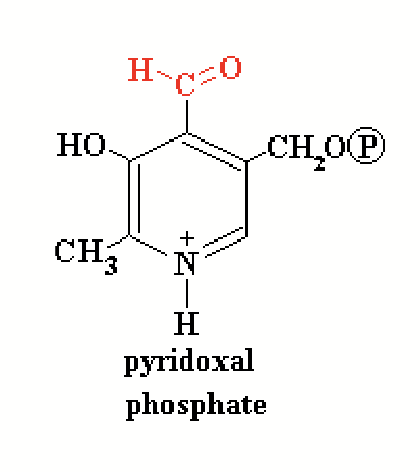
prosthetic group
A non-amino acid helper molecule that an enzyme may contain as part of its structure. It binds at the enzyme catalytic site and initiates reaction by withdrawing electrons from the substrate.
general acid catalysis
Catalysis by an amino acid side chain that donates H+ to the reaction.
H+ exchange takes place right at the site of reaction, so the pH of surroundings is not affected (as enzymes must function at physiological pH)
Gain or loss of one H+ in a small confined volume can have the same effect as strong acid or base
general base catalysis
Catalysis by an amino acid side chain that removes H+ from the reaction.
H+ exchange takes place right at the site of reaction, so the pH of surroundings is not affected (as enzymes must function at physiological pH)
Gain or loss of one H+ in a small confined volume can have the same effect as strong acid or base
chemical catalysis
Nucleophilic catalysis
Electrophilic catalysis
General acid catalysis
General base catalysis
These mechanisms can each contribute about an 100-fold increase in reaction rate, and work together to lower Ea.
Some reaction mechanisms use multiple of these!
stabilizing the transition state
Reactions must pass through a transition state to proceed, and key atoms may change shape (e.g. trigonal planar → tetrahedral), or a bond may stretch.
Less activation energy is needed if the enzyme active site is complementary to the transition state (the transition state differs chemically from the substrate).
The enzyme can help by binding the substrate in the ideal shape for the transition state (catalytic site set up to complement the transition state of the substrate, thus lowering energy content).
transition state
What reactions must pass through to proceed. Key atoms may change shape (e.g. trigonal planar → tetrahedral) or a bond may stretch. This state differs chemically from the substrate. If the active site is complementary, less activation energy is needed.
Activation energy can be lowered partly be choosing a different state, and partly by making the enzyme contribute to bond distortion that leads to this state.
enzyme assay
The process of measuring enzyme-catalyzed reaction rate.
Enzymes speed up reaction rate in proportion to the amount of enzyme present!
enzyme kinetics
The mathematical analysis of how rate varies as a function of substrate concentration: can be used to test reaction mechanisms.
Measure rates based on the rate of disappearance of reactant or rate of appearance of product
Enzymes speed up reaction rate in proportion to the amount of enzyme present!
artificial substrate
A molecular “look-alike” for a real substrate. Often used in trypsin reactions to increase efficiency.
Trypsin hydrolyzes peptide bonds on the C-terminal side of Arg or Lys except when Pro is present in that position → we hydrolyze peptidyl nitroanilide, which releases p-nitroaniline which is distinctly coloured and thus makes it easy to measure the concentration and the rate.
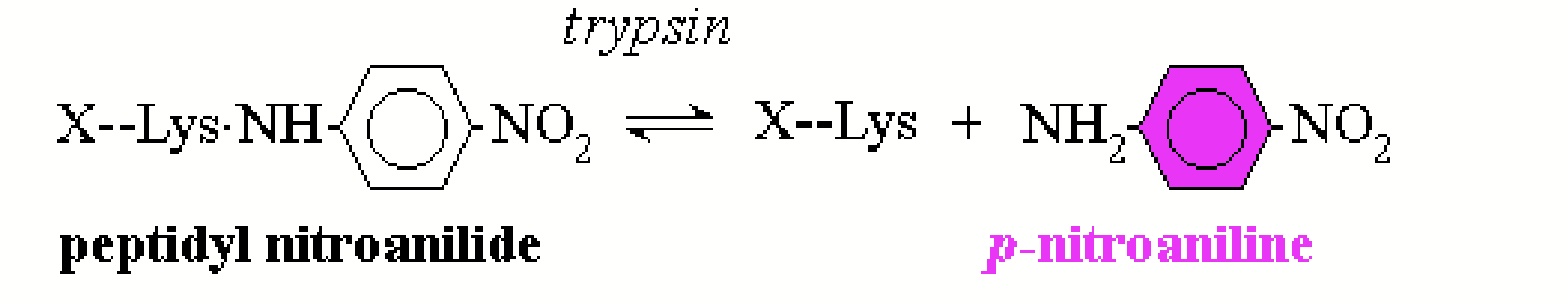
UV absorbance change
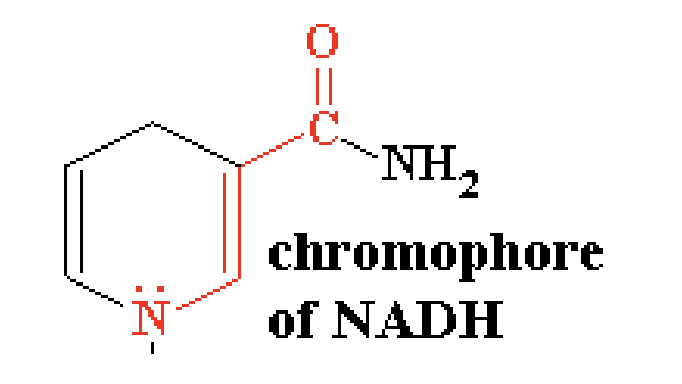
Some natural substrates show UV absorbance change after conversion to product.
When pyruvate is converted to lactate, it converts NADH to NAD+ (oxidated). NADH absorbs ultraviolet light at 340 nm, whereas the NAD+ product does not absorb → Overall absorbance decreases as the reaction proceeds ([NADH] decrerases as it is converted to NAD+, thus, less absorbance).
![<p>Some natural substrates show UV absorbance change after conversion to product.</p><p>When pyruvate is converted to lactate, it converts NADH to NAD<sup>+ </sup>(oxidated). <strong>NADH </strong>absorbs ultraviolet light at 340 nm, whereas the NAD<sup>+</sup> product does not absorb → Overall absorbance decreases as the reaction proceeds ([NADH] decrerases as it is converted to NAD<sup>+</sup>, thus, less absorbance).</p>](https://knowt-user-attachments.s3.amazonaws.com/fb704663-92fe-48ec-9d81-bc4bbf3c716b.png)
chromophores
Parts of molecules with conjugated double bonds (alternating double and single bonds). Absorb visible or UV light.
Coloured compounds absorb between 400-700 nm
Natural biochemical forms frequently absorb in the UV range, 200-400 nm
Larger forms absorb at longer wavelengths
conjugated double bonds
Alternating double and single bonds.
Beer-Lambert Law
Absorbance is proportional to sample concentration and sample thickness.
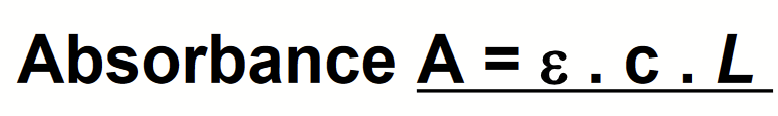
Absorbance
Io = initial intensity
I = measured intensity

rate of reaction
Quantitative description of enzyme catalysis
Change in [substrate] or [product] per unit time (Mmin-1 or molL-1min-1).
= [substrate used] / time or [product formed] / time (in M or mM)
enzyme activity
Quantitative description of enzyme catalysis
Moles of substrate converted per unit time (µmol/min). Represents the quantity of enzyme present.
= rate * volume
Also: (moles substrate or moles product) / time
specific activity
Quantitative description of enzyme catalysis
The number of enzyme units per milligram of total protein (µmol min-1 mg-1 or µmol min-1 µg-1).
Measure of enzyme purity during purification of enzymes and measure of enzyme efficiency when two different pure enzymes are compared.
= (Enzyme Activity) / (Total Protein)
molar activity
Quantitative description of enzyme catalysis
Activity per mole of enzyme (min-1).
Equal to the turnover number, the number of catalytic reaction cycles per molecule of enzyme per second.
= (Specific Activity)(Molar mass of enzyme) → with necessary unit conversions!
If n moles of substrate are converted per second per mole of enzyme, then n molecules of substrate are converted per molecule of enzyme.
kinetics
The mathematical analysis of how reaction rate varies as a function of reactant concentration.
progress curve
Plot substrate or product concentration over period of time ([S]).
Initial rate vo is taken from the slope of the curve at time zero.
Measure several rates at different initial [S].
![<p>Plot substrate or product concentration over period of time ([S]).</p><p>Initial rate v<sub>o</sub> is taken from the slope of the curve at time zero.</p><p>Measure several rates at different initial [S].</p>](https://knowt-user-attachments.s3.amazonaws.com/a7ca2497-47c7-408c-b09e-02f619e08f3a.png)
normal chemical reactions
Follow simple rate laws.
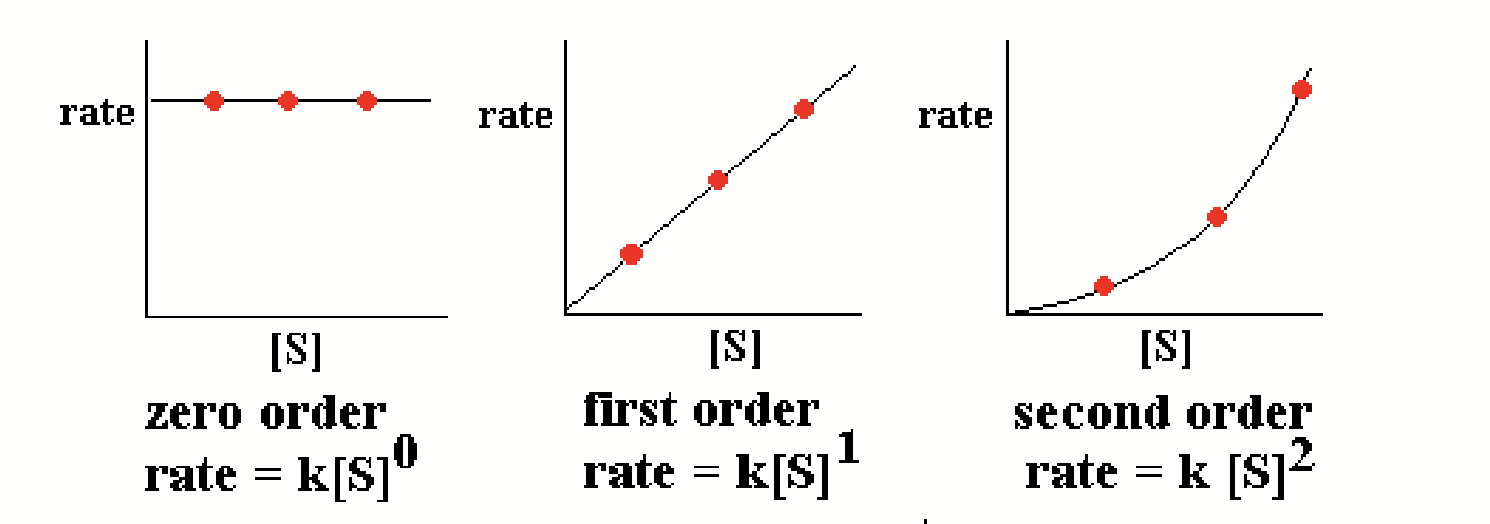
enzyme reaction
Does not follow a simple rate law - instead, creates a hyperbolic curve that plateaus as [S] increases.
![<p>Does <strong>not</strong> follow a simple rate law - instead, creates a <strong>hyperbolic curve</strong> that plateaus as [S] increases.</p>](https://knowt-user-attachments.s3.amazonaws.com/b665f0e0-8074-4272-9ea8-992f96a1c5db.png)
analysis of enzyme reaction as two steps
Since enzyme is recycled, it is not consumed; [E]total is constant.
Work at time = 0 so [P] = 0, reverse reaction at step 2 can be ignored.
Individual steps have rate constant k1, k-1, k2.
![<p>Since enzyme is recycled, it is not consumed; <strong>[E]<sub>total</sub></strong> <strong>is constant.</strong></p><p>Work at <strong>time = 0</strong> so <strong>[P] = 0</strong>, <strong>reverse reaction at step 2 can be ignored</strong>.</p><p>Individual steps have <strong>rate constant k<sub>1</sub>, k<sub>-1</sub>, k<sub>2</sub>.</strong></p>](https://knowt-user-attachments.s3.amazonaws.com/7e408cbf-febf-491d-b1cb-cdb8043976f0.png)
Michaelis-Menten equation
Equation that shows how initial rate (vo) varies as a function of substrate concentration ([S]). May also be written in the fractional form, which is useful for solving enzyme kinetics problems.
Every enzyme has characteristic values of the two constants, Vmax and KM.
![<p>Equation that shows how <strong>initial rate (v<sub>o</sub>)</strong> varies as a <strong>function of substrate concentration ([S])</strong>. May also be written in the fractional form, which is useful for solving enzyme kinetics problems.</p><p>Every enzyme has characteristic values of the two constants, <strong>V<sub>max</sub></strong> and <strong>K<sub>M</sub></strong>.</p>](https://knowt-user-attachments.s3.amazonaws.com/bfbe4767-060c-4211-a068-0d6b70264690.png)
Vmax
The upper limit for rate; indicates the catalytic rate when 100% of the enzyme is occupied by substrate (saturated with substrate). Higher rate means a faster reaction and better catalysis.
At high [S], all enzyme molecules have bound substrate and are engaged in catalytic activity, so reaction cannot go any faster (adding substrate cannot increase rate).
Considered a pseudo-constant (constant only if the amount of enzyme is fixed).
turnover number (k2)
The rate that a singular molecule is working; how fast a single enzyme molecule can work. The true constant!
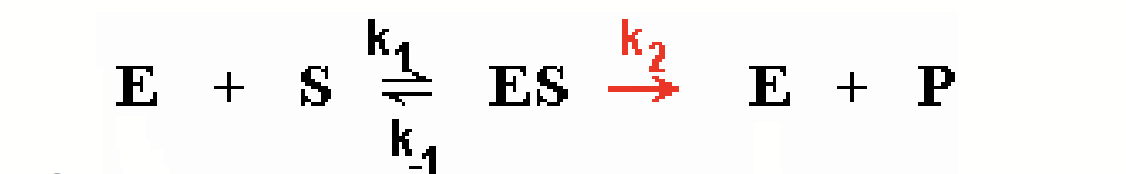
Michaelis constant (KM)
Indicates how well the substrate fits the catalytic site.
The concentration of substrate [S] at which vo is equal to 50% of the maximum rate Vmax.
A low value indicates that the enzyme binds and utilizes substrate well; less [S] is needed to occupy the enzyme.
A high value indicates that the enzyme binds and utilizes substrate poorly; more [S] is needed to occupy the enzyme.
An enzyme with more than one substrate has a different value for each.
Lineweaver-Burk method
Method using linear transformations to convert the Michaelis-Menten equation into a straight line form; slopes and intercepts of the straight line give better estimates of Vmax and KM. Work well unless there are significant errors in the data.
Take reciprocals of both sides of the Michaelis-Menten equation; obtain KM by extending the graph onto the negative x-axis
Why do this? Real experimental data often shows scatter due to measurement errors
inactivators
Usually react with enzymes irreversibly. Results from covalent chemical reactions. Reaction destroys catalytic activity and “uses up” the enzyme.
Many are highly toxic (e.g. nerve gases).
Simple stoichiometric relationship!
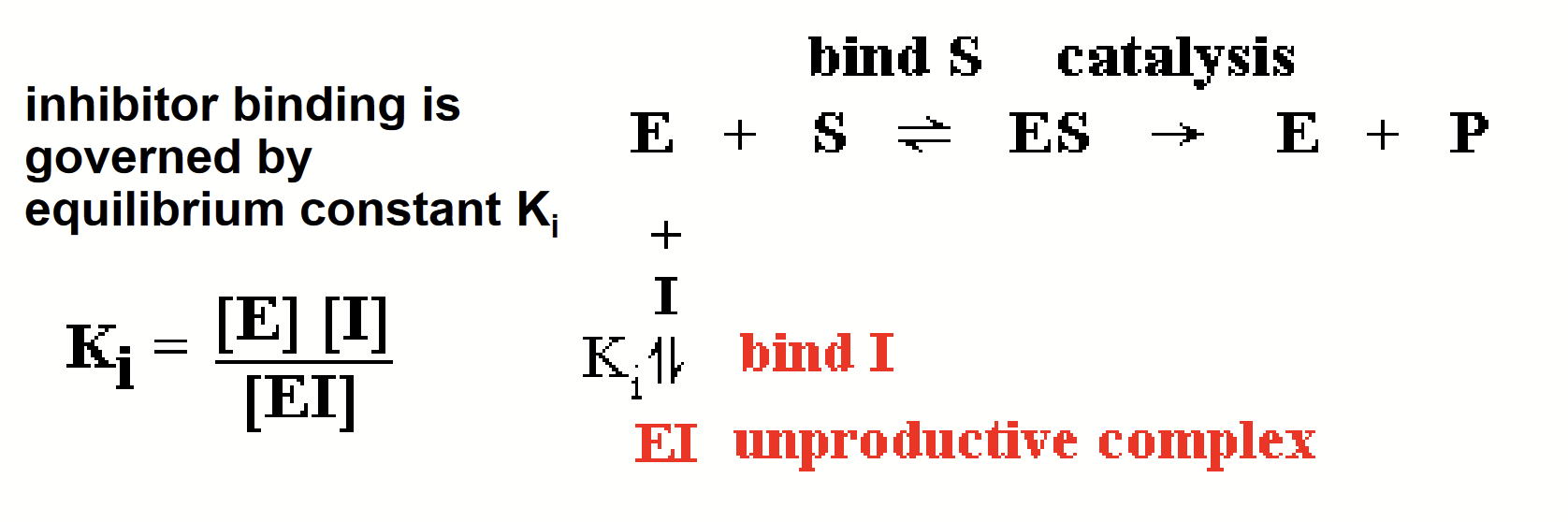
inhibitors
Decreases enzyme activity without destroying the catalytic function of enzyme molecule, usually bind to enzymes reversibly. Enzyme activity is restored if concentration is reduced.
Binds to site on the enzyme by non-covalent forces, similar to substrate (H-bonding, hydrophobic effect, etc.).
Degree of inhibition is governed by binding equilibrium, not simple stoichiometry.
Presence may affect different stages of the catalytic reaction, gives different modes of inhibition.
Can regulate enzyme activity in the cell → more economical for a cell to make and destroy a small inhibitor than a large enzyme.
modes of inhibition
The presence of inhibitor may affect different stages of the catalytic reaction, giving different modes.
Competitive: affects ability to bind substrate
Non-competitive: affects catalytic rate (also mixed)
competitive inhibition
Affects the ability to bind substrate.
Arises when inhibitor can only bind to unoccupied enzyme E.
Formation of EI complex means less available E available to bind substrate. Inhibitor and substrate compete for available enzyme - high [S] can overcome the inhibitor (in this case, can still reach Vmax).
S and I often share the same binding site and may resemble one another in terms of chemical structure.
non-competitive inhibition
Affects the catalytic rate (also mixed inhibition).
Formation of EI and EIS means less ES to undergo catalysis, but substrate can still bind to EI without yielding product.
Inhibitor binding site is different from substrate binding site.
Bound inhibitor may disorganize the catalytic component of the enzyme.
If EI and EIS steps each have a different Ki, we get mixed inhibition.
regulation of enzyme activity
Can regulate enzyme activity in the cell → more economical for a cell to make and destroy a small inhibitor than a large enzyme.
e.g. aspirin, ibuprofen
equilibrium constant Ki
Constant that governs inhibitor binding.
competitive inhibition Lineweaver-Burk plot
Different lines represent different [I]
No effect on Vmax, so all graph lines have the same y-intercept (y-intercept = 1/Vmax)
KM’ increases as [I] increases, so x-intercept gets smaller when considering absolute values (x-intercept = -1/KM)
Slope increases as [I] increases (slope = KM’/Vmax)
![<ul><li><p>Different lines represent different [I]</p></li><li><p>No effect on V<sub>max</sub>, so all graph lines have the same y-intercept (y-intercept = 1/V<sub>max</sub>)</p></li><li><p>K<sub>M</sub>’ increases as [I] increases, so x-intercept gets smaller <strong>when considering absolute values</strong> (x-intercept = -1/K<sub>M</sub>)</p></li><li><p>Slope increases as [I] increases (slope = K<sub>M</sub>’/V<sub>max</sub>)</p><p></p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/53f4b508-52ac-49bb-84bc-1270e1e07dac.png)
non-competitive inhibition Lineweaver-Burk plot
Different lines represent different [I]
No effect on KM, so all graph lines have the same x-intercept (x-intercept = -1/KM)
V’max decreases as [I] increases, so y-intercept gets bigger (y-intercept = 1/V’max)
Slope increases as [I] increases (slope = KM/V’max)
For mixed, lines meet above the x-axis
![<ul><li><p>Different lines represent different [I]</p></li><li><p>No effect on K<sub>M</sub>, so all graph lines have the same x-intercept (x-intercept = -1/K<sub>M</sub>)</p></li><li><p>V’<sub>max</sub> decreases as [I] increases, so y-intercept gets bigger (y-intercept = 1/V’<sub>max</sub>)</p></li><li><p>Slope increases as [I] increases (slope = K<sub>M</sub>/V’<sub>max</sub>)</p></li><li><p>For mixed, lines meet above the x-axis</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/999ec5f4-eecf-41d9-8f19-2e8c5367812a.png)