BIOL 200
1/274
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
275 Terms
What is life? (6 things)
high degree of chemical and structural complexity
a capacity to extract, transform, and use energy from the environment
Defined functions for the organism’s components and regulated interactions among them
a capacity to sense and respond to environment
a capacity to self-replicate
a capacity to evolve
Life is built by
complex macromolecules
to function, life requires large organic molecules (and structures formed by them)
Without energy, molecules
decay toward more disordered states and to come to equilibrium with its surroundings
what does life require from the environment?
continual imput of energy
cells are open/closed systems
open
what is an open system
they require the import or energy sources and raw materials, and export of waste
lipid membrane
selective barrier
greater concentration of nutrients and synthesized products inside the cells
allow cells to have a distinct intracellular environment very different from its surroundings
to be outside of a chemical equilibrium with its surroundings
Most molecular function is accomplished by
proteins
each protein serves a specific function
Proteins serve (3 things)
structural roles
generate movement
sense and detect signals
Many proteins are capable of
catalyzing reactions (enzymes) including those needed to obtain and use energy from the environment
Genetic information is stored in
DNA
genetic information = information to self-replicate
DNA (and RNA) have __ building block
4 (nucleotides), making them much simpler than proteins
proteins have __ building blocks
20 (amino acids)
Central Dogma of Molecular Biology
DNA contains information to make proteins
DNA → RNA → proteins
cells cannot go in the opposite direction of this flow
DNA is also required as a template to synthesize DNA
First Step of Making Protein
Genetic information is copied to a third type of molecule, RNA
Process is called Transcription
mRNA
messenger RNA is the result of transcription
exported from the nucleus and used by large molecular machine called ribosome
ribosomes
synthesize proteins using the information in RNA
called translation
translation
nucleotides are read in triplets (codons)
greater information is conveyed in RNA (4³ = 64 combinations)
requires two other types of RNA (tRNA and rRNA)
tRNA
transfer RNA
coupled into an amino acid residue, pairs with the sequence in mRNA during translation to convert the sequence in codons into specific amino acids
rRNA
ribosomal RNA
make up ribosomes
composed of proteins and RNA
catalyze protein synthesis
catalytic side that synthesizes is the rRNA part
Three important polymers in molecular biology (macromolecules)
DNA
RNA
Proteins
information in cells is carried by polymers
polymer
large molecules composed of multiple covalently linked building blocks (small molecules) called monomers
ex. PET, most common type of polyester
monomer is ethylene terephthalate
How to cook PET
requires mixing two precursors
input of energy, in the form of heat (150-290 C)
removal of byproducts (such as methanol)
proteins are formed by amino acids
monomers that form proteins are the amino acids
amino acid structure
central carbon atom (alpha carbon), an amino group, a carboxyl group, and a side chain (R group)
polymerization of amino acids
amino acids are joint together by the reaction of carboxyl group in one of them with the amino group of a second amino acid
reaction results in the formation of a peptide bond
a polymer of amino acids is called
polypeptide
backbone of protein
the linear chain of carbon and nitrogen atoms that contain the peptide bonds
side groups stick out of the backbone
protein orientation
an amino group at one end (N-terminus) and a carboxyl group at the other (C-terminus)
Side chains
20 different side chains = 20 different amino acids
differ in their size, shape, charge, hydrophobicity, and chemical reactivity
essential amino acids
the 9 of the 20 amino acids humans and other mammals can’t synthesize and must consume in diet
3 groups of amino acids
hydrophobic, hydrophylic, and special amino acids
hydrophobic amino acids
poorly soluble in water
contain linear or branched hydrocarbons or contain aromatic rings
hydrophylic amino acids
readily soluble in water
can either be positively (basic), negatively (acidic), or polar with uncharged groups
special amino acids
cysteine: reactive sulfhydryl group (SH)
serves in catalysis of enzymes
reacts with other cysteins to form disulphide bonds to join two polypeptides
glycine: smallest amino acid
fits in small spaces
side chain H
proline: side chain that forms a covalent bond to the N in th eamino group attached to the alpha carbon
incorporation of proline into proteins generates kinks in the linear chain
DNA and RNA are formed by
nucleotides
the monomer of nucleic acids (DNA and RNA) are called nucleotides
nucleotides
composed of pentose (5-C sugar), a phosphate group, and a ring-shaped molecule containing nitrogen called nitrogenous base
negative charge in phosphate makes them acidic
DNA + RNA have an overal -ve charge
pentose in RNA and DNA
the pentose in RNA is ribose
the pentose in DNA is 2’-deoxyribose
the difference is the loss of an OH group at position 2’ in deoxyribose
RNA and DNA use 4 bases each
purines: pair of fused rings
pyrimidines: contain a single ring
Thymine is only present in
DNA
Uracyl is only present in
RNA
Bases are connected to the ribose through
the nitrogen atom at position 9 (in purines) or 1 (in pyrimidines)
nucleotides have a ribophosphate backbone
nucleotides are joined through two phosphodiester covalent bonds where a phosphate group links two (deoxy)ribose at positions 5’ and 3’
phosphate and (deoxy)ribose groups generate the backbone of nucleic acids
DNA and RNA have an orientation with a 5’ end (containing a phosphate) and a 3’ end (containing a hydroxyl group)
each monomer can contain a different base, therefore generating a unique sequence
RNA is more prone to breaking than DNA
hydroxyl group at position 2’ in the ribose of RNA can spontaneously react with the phosphate group, break the RNA molecule
DNA lacks an OH at the 2’ of ribose, making it much more stable and the choice for the genetic material in cells
some viruses have genomes of RNA
DNA molecules in our genome are hundreds of millions of base pairs in length
Purines and Pyrimidines pair in DNA
the purine Adenine (A) pairs with the pyrimidine Thymine (T)
kept together by 2 hydrogen bonds (weak non-covalent bonds)
the purine Guanine (G) pairs with the pyrimidine Cytosine (C)
interact through 3 hydrogen bonds
DNA is double stranded
in cells, two DNA polymers ‘anneal’ (pair) to form a double-stranded following the pairing of bases (A-T and G-C)
the sequence of bases in the two strands is complementary
pairing between the two strands is antiparallel
individually weak, the sum of many H-bonds help keep the two strands together
double helix structure
ribophosphate backbone in the outside and the bases inside
bases are regularly spaced 0.34 nm appart
on the outside of the helix, the spaces between the strands form two helical grooves of different widths: the major and minor grooves. Atoms on the edges of each base are exposed
Double-stranded DNA (dsDNA) shapes into a
right-handed double helix
Some proteins recognize specific DNA sequence motifs
structure of DNA allows DNA-binding proteins to “read” the sequence in it without unwinding double helix
major groove is wide and contains unique chemical information
most protein-DNA interactions use the major-groove
minor groove is shallow and cannot distinguish all bases
Evidence of Common ancestry
shared use of amino acids and nucleotides across life
the use of only L amino acid stereoisomers to form proteins
conservation of the codons that code for the same 20 amino acids
machinery for protein synthesis is similar across organisms
catalytic site of ribosomes is in the rRNA
protein function is determined by…
their structure
hierarchy of protein structure
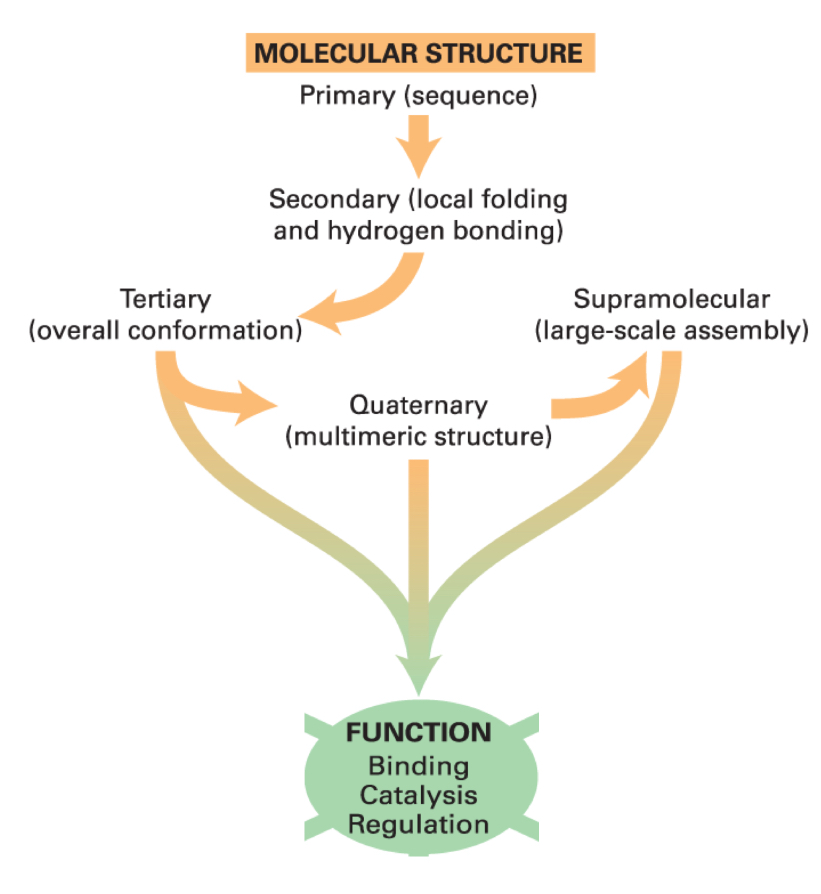
what folds into a specifc 3D structure to acquire a function?
linear polypeptide chains
the properties of amino acids direct the folding to specific structures
primary structure (sequence)
the sequence of a protein has a direction, with a beginning (N-terminus) and an end (C-terminus)
properties are given by the side chains
number of amino acid residues and their mass in daltons is what protein size is expressed as
smallest proteins are ~40 amino acids in length. the “average” protein depends on the organism. In yeast, the average is 466 aa residues
largest protein has 34,000 aa residues
oligopeptides
small chains <30 amino acids
polypeptides
larger chains, single chain, often 200-300 amino acids
amino acid residue
an amino acid that has been incorporated into a peptide or protein chain through a dehydration reaction
green flurescent protein example
238 aa in length
26.9 kDa
absorbs blue light and transforms it into green light
secondary structure
local conformations of the peptide chain backbone, generating stable arrangements of the aa residues
two major peptide chain backbone conformations
alpha-helix
beta-sheet
both are based on H-bonding (stabilizes) between peptide bond carbonyl O atoms and one amino acid residue, and amide H atoms on a different amino acid residue
~60% of the length of the average polypeptide chain consists of segments of alpha-helix and beta-sheet. multiple secondary structure elements per polypeptide
structure of alpha-helix
don’t contain proline
side chains can be hydrophobic, hydrophilic, or charged
H-bonds occur between AA in same chain (C=O and N-H)
happens in the position n+4
tilted axis generates a periodicity of 3.6 aa residues per turn
backbone has a straight rod structure with side chains pointing outward
surface properties depend on side chains. so does the alpha-helix formation and its interactions with other parts of the protein and other molecules
structure of beta-strand
short (5-8 aa)
H-bonds formed between two adjacent Beta-strands oriented perpendicularly to the chains of the backbone
alignment of 2 or more beta strands generate nearly 2-D sheets
side chains protrude above and below the beta-sheet plane. they determine the interactions within the protein and with other molecules, and the propensity to form
beta-strand organization
B-strands in the same polypeptide can be parallel or antiparallel
B-strands from two different proteins can also interact to form sheets
motif
combination of two or more secondary structures that form a distinct 3D structure found in multiple proteins
associated to a specific function
tertiary structure
the overall conformation of the polypeptide
the spatial organization of the multiple secondary structure elements
stabilized by hydrophobic interactions (most important one), van der walls, and Hbonds
forces stabilizing the tertiary structure are weak, resulting in fluctuations in the structure
structure acquisition is partly driven by
the effect of hydrophobicity
water molecules surrounding hydrophobic molecules dispersed in water adopt a constrained, cage-like organization (low entropy)
if hydrophobic molecules coalesce, the total number of low entropy, constrained water molecules is reduced, and this net increase in entropy ultimately drives the formation of separate hydrophobic and aqueous phases
coalescence of hydrophobic molecules also favoured by weak non-covalent van der Waals intermolecular interactions
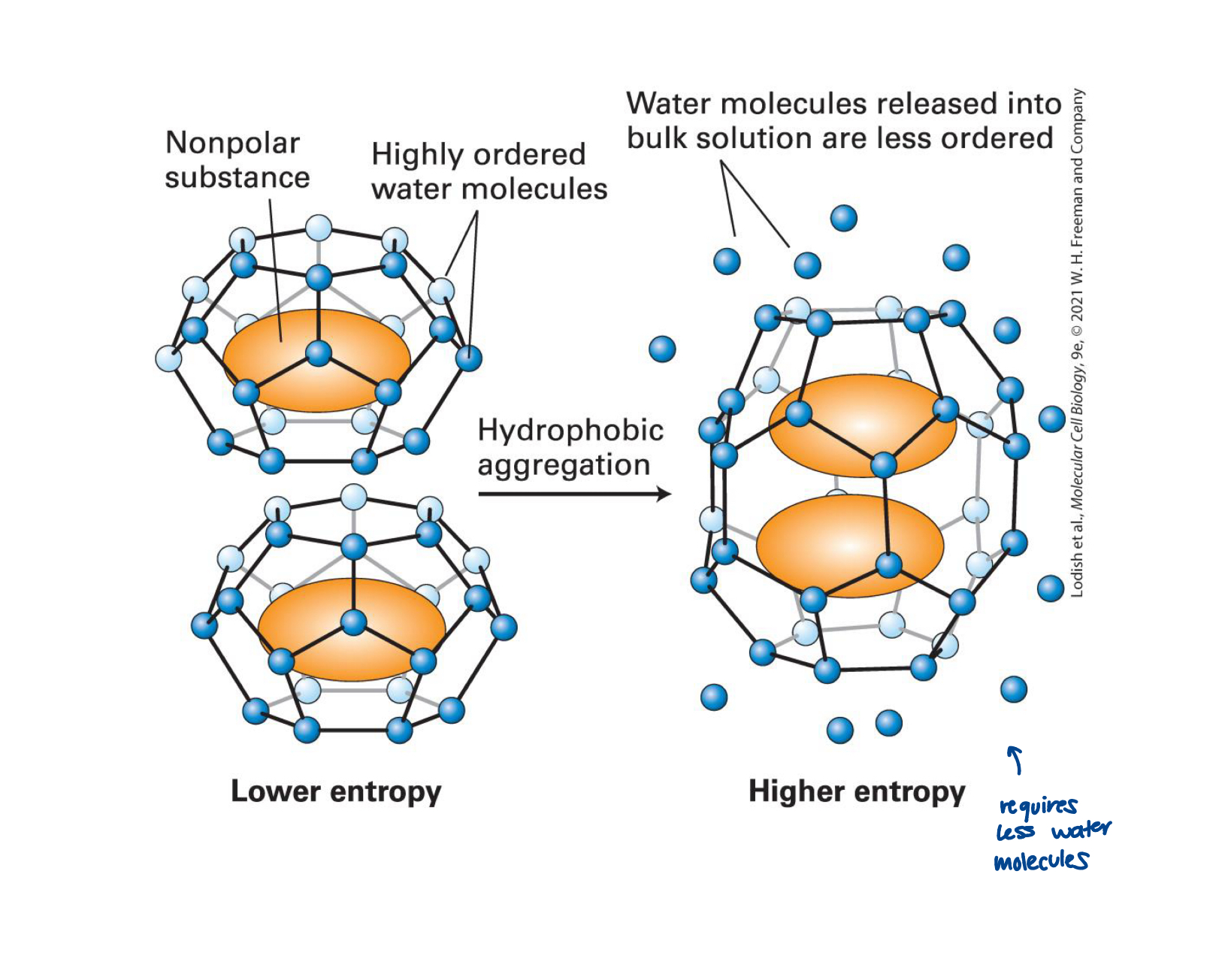
oil drop model of protein folding
hydrophobic and hydrophylic side chains are distributed throughout the linear sequence
when the protein folds, the hydrophylic side chains tend to be exposed to water and hydrophobic residues tend to cluster together in the inner core, hiding form the aqueous surroundings
conformations
each protein adopts only one or a small number of similar structures
domains
distinct regions of the proteins structure
how tertiary structures are divided
they can represent a particular funciton, structure, or refer to the spatial relationship with the rest of the protein
functional domains can be seperated from the rest of the protein and remain functional
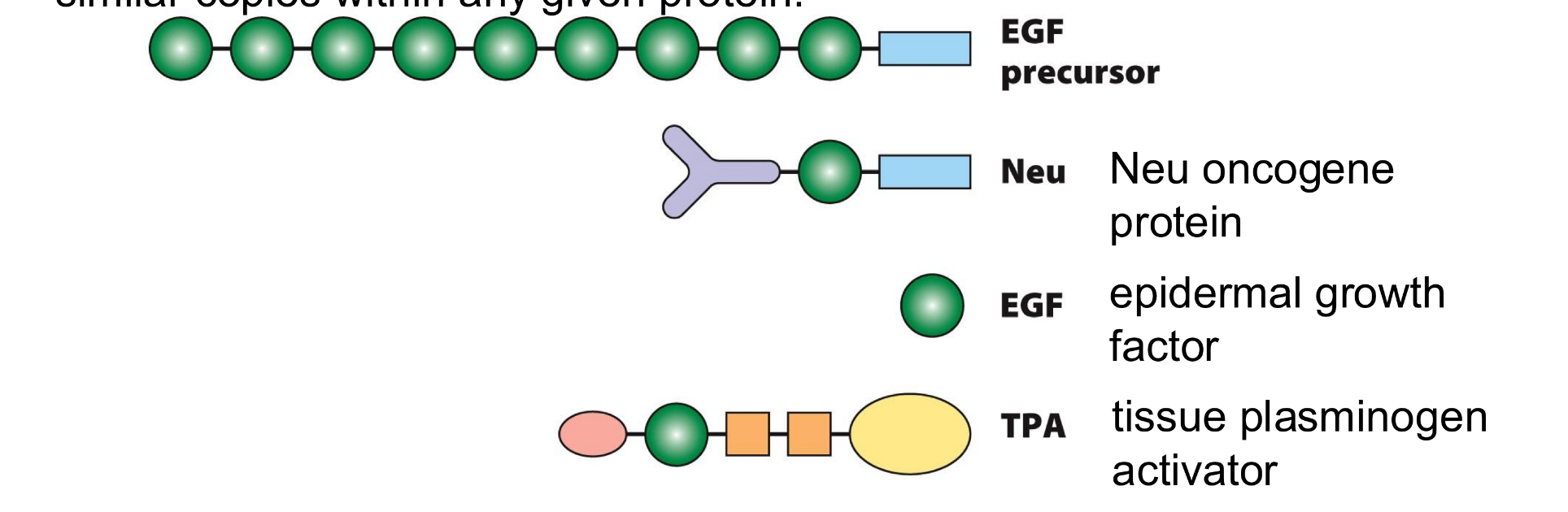
modular nature of proteins
similar domains can be found in diverse proteins, and also as multiple similar copies within any given protein
shapes/colours show a variety of distinct domain types, including EGF domain (green) and membrane-spanning domain (blue)
tertiary structure of GFP (green fluorescent protein)
explains activity
the typical structure of FP consist of a barrel containing 11 beta sheets and central helix that contains the chromiphore
interior of the barrel is highly crowded by side-chains of amino acid residues, limiting movement inside
alpha-helices at the ends of the protein serve to stabilize it
chromophore is formed by a post-translational modification that result in the cyclization of peptide backbone
quaternary structure
multimeric proteins can contain any number of identical or different polypeptides
supramolecular complexes
large “molecular machines” made up of multiple distinct proteins, each of which may itself contain multiple subunits
ex. transcription initiation complex
techniques to determine protein structure
X-ray crystallography and cyroelectron microscopy
both require the purification of individual proteins
generally representing months or years of work
motifs and domains
each domain can contain multiple motifs
unstructured regions
regions in DnaA without a fixed structure (more flexible). This is the case for most proteins.
DnaA
a bacterial protein that initiates DNA replication
contains 4 domains
the domain IV contains two motifs that allow the interaction with specific sequences on DNA:
A Helix-Turn-Helix and a Basic Loop
the domain III contains motifs for the binding and hydrolysis of ATP:
Walker A and Walker B
it contains also other motifs for multiple other functions
motif
the combination of two or more secondary structures that form a distinct 3D structure found in multiple proteins. Associated to a particular function
protein folding and hierarchy of protein structure
polypeptide first folds regions with secondary structure
secondary structure elements group into motifs then into domains
then the tertiary structure is acquired
each amino acid residue can rotate on its axis
at aa residue position of the polypeptide, the axis of the backbone can rotate at the bonds connecting the alpha carbon to the carbonyl and aminde groups
rotation is limited due to steric constrains imposed by the backbone and the side chains
proline confers special conformation to proteins
most amino acid residues in the polypeptide chain orient in a trans configuration relative to the peptide bond, resulting in a linear backbone
in contrast, between 5-7 percent of the peptide bonds with proline acquire a cis-configuration
proline helps direct protein folding
isomerization between trans and cis configurations can occur spontaneously, but it is slow
peptidyl-proline isomerases (PPlases) are enzymes that speed up the process during folding
isomerization of a single proline can dramatically change protein structure, and affect its activity
how long does folding take
micro- to milliseconds
why can proteins fold when isolated in test tube
because the information required for the acquisition of structure is in the sequence
despite the limitations in rotation at each residue, the combination of many small changes in the orientation of proteins allow them to twist and turn
where are the hydrophobic aa residues
inside the correctly folded proteins
hydrophobic patches at the surface of a protein is a sign of misfolding
chaperones
proteins that help guide protein folding along productive pathways, by permitting paritally misfiled proteins to return to the proper folding pathway
recognize exposed hydrophobic residues
upregulated in conditions where misfolding is increased (ex. heat-shock)
can fold newly made proteins, refold misfolded or unfolded proteins, and disassemble potentially toxic protein aggregates that form due to protein misfolding
work through ATP-dependent cycles of binding to, and release from, misfolded “client” molecules, at exposed hydrophobic patches. by blocking the exposed hydrophobic patches the chaperaone keeps the folding or refolding protein isolated, while productive folding events occur
two types: chaperones and chaperonins
ex. Hsp70 (Heat-shock protein)
→ binds to short segments of an unfolded proteins such as those newly synthesized as they emerge from the ribosome
→ binding and hydrolysis of ATP results in changes in the conformation of Hsp70 and are needed for its function
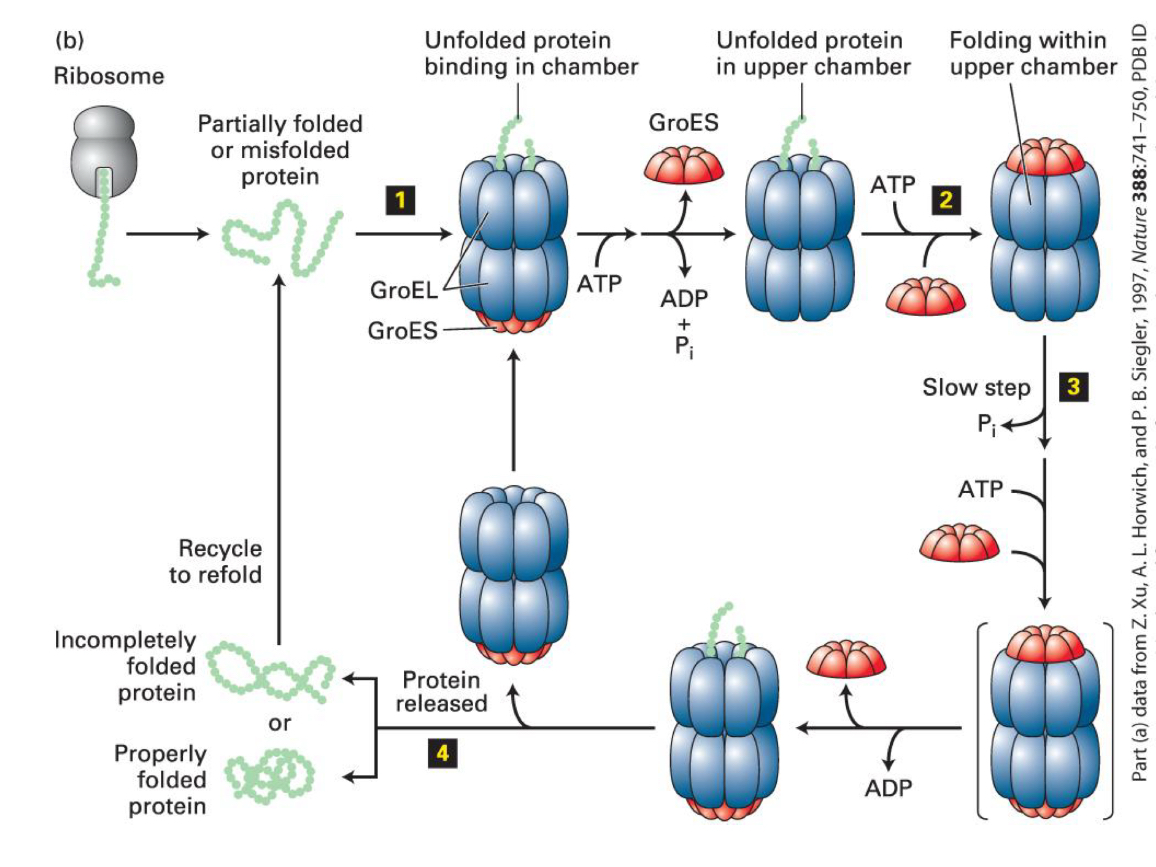
chaperonins
large complexes that isolate unfolded proteins
ex. GroEL is composed by two stacked rings, each composed of seven subunits
→ each ring interacts with a seven-subunit co-chaperone that acts like a lid, GroES, also containing seven subunits
→ in the center of GroEL there are chambers where all or part of a protein enters
chaperonin mode of action
proteins of less than 60 kDa in mass are captured by hydrophobic residues near the entrnce of the GroEL chamber
ATP hydrolysis regulates the cycle
protein folding in the two rings is coordianted
multiple cycles may be requried for proper folding
what does degredation target?
irretrievably misfolded proteins
protein degradation
ubiquitin/proeasome system
poly-ubiquitin “tags” damaged or misfolded proteins for degradation
ubiquitin-tagged proteins are fed into a multi-subunit chamber in which the subunits form inward-facing proteases
ubiquitylation
important protein regulator
small ubiquitin protein (76 aa residues, 8.6 KDa) becomes covalently linked to the lysine residue of target proteins
ubiquitin conjugating system
carboxyl terminus is activated
ubiquitin is transferred to a ubiquitin conjugating subunit (E3 ubiquitin ligase)
600 E3 coded in our genome, each with specific substrate binding
polyubiquitinylation
marks protein for degradation
ubiquitin ligases recognising exposed hydrophobic reisudes add multiple ubiquitins to proteins, forming chains of 4 or more ubiquitins
polyubiquitinylated proteins are recognized by Ub receptors in the proteasome
Deubiquitinases (Dubs) hydrolyze bonds between ubiquitous to recycle them
ATPase driven auxillary proteins unfold proteins and transports to the core for degradation
once inside the inner chamber, polypeptides are digested into short fragments of 2-24 aa in length
peptide bonds of hydrophobic, acidic, and basic residues are cleaved at the active site
the resulting polypeptides are further degraded into single amino acids in the cytoplasm
what leads to protein aggregation?
accumulation of misfolded proteins
due to the imperfect system of refolding with chaperone assistance or multiubiquitination/proteasome mechanism
protein aggregation
misfolded proteins or incompletely degraded proteins can interact with each other, hiding their hydrophobic residues, forming aggregates
high protein concentration and changes in environmental conditions leads to formation of aggregates
can be amorphous or well-organized, such as in the amyloid state
amyloid fibrils
formed by the generation of short segments (6-12 residues in length) that form long arrays, or filaments, of B-sheets
each B-strand is oriented nearly perpendicularly to the axis of the filament. two stacks stwist about one another, forming protofilaments, many of them forming fibrils
amyloids of makers for disease, why?
found in tissue
neurodegenerative diseases (ex. alzheimers and parkinsons) all contain amyloids
amyloid formation is associated to age, but it is also more prevalent in mutant proteins
Binding
diverse functions performed by proteins are based on their ability to engage in binding
proteins bind to one another, to other macromolecules, to small molecules, and to ions
ligand
the molecule to which a protein binds
specificity
important to protein binding
the ability of a protein to bind to only one particular ligand, even in the presence of a vast excess of irrelevant molecules