Theme 2 Modules 1 & 2 Review Lecture Bio1A03 - Copy (2)
1/43
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
44 Terms
How are the processes different in time and locations in prokaryotes compared to eukaryotes?
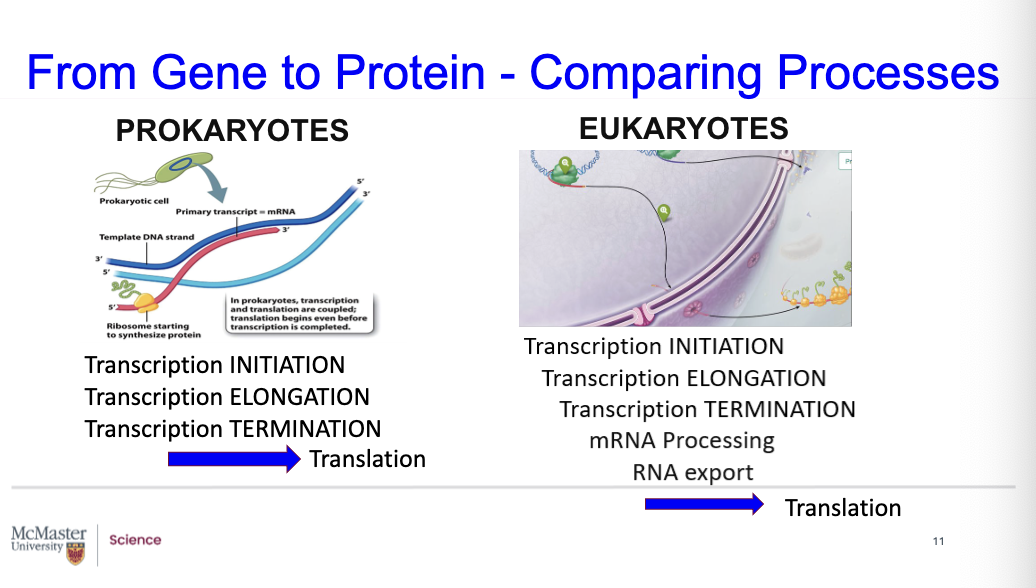
With prokaryotes, such as Archaea & bacteria, there are 3 phases of transcription: 1. Initiation, 2. Elongation, 3. Termination.
These are not separated in location, since in prokaryotes, the DNA is not enclosed in a membrane-bounded nucleus. The DNA is in the nucleoid region. You can have both transcription and translation coupled. These processes can occur simultaneously.
With the eukaryotes, there are also the phases 1. Initiation, 2. Elongation, 3. Termination. But there is the separation in time and in location. Transcription occurs within the nuclear envelope. There is also mRNA processing that occurs, and mRNA export out of the nucleus, through the nuclear pore complex, to a separate location where translation occurs, as shown in this figure in the cytoplasm at the position of the ribosomes.
Dna cant go outslide the nuclear envelope but mRNA can
MRNA venture out ot eh nucelar envelope to go to the ribisomes
Home Study Task - make a study table that compares transcription & translation in prokaryotes and eukaryotes.
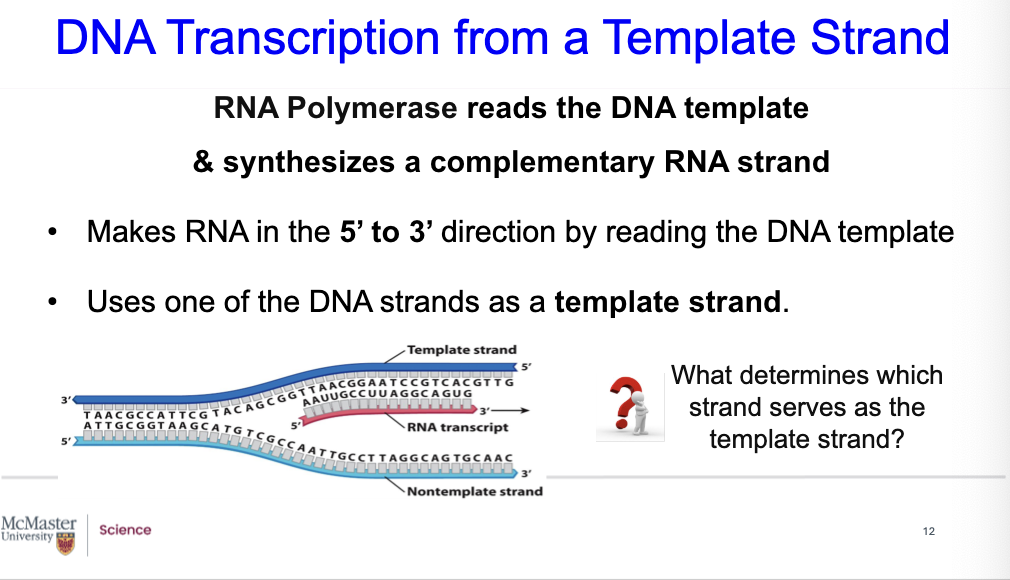
who discovered RNA polymerase? What determines which strand serves as the template strand?
Looking at it from the template strand, we see RNA polymerase (enzyme), which was discovered in the 1960’s by Weiss & coworkers. It reads the DNA template and synthesizes a complementary RNA strand.
It makes RNA in the 5’ to 3’ direction by reading the DNA template.
It uses one of the DNA strands, as a template strand.
What determines which strand serves as the template strand?
It depends on the orientation of the promoter and on other regulatory molecules.
Researchers are still investigating this.
different name for template strand and non template strand
Template, anti sense, non coding
Non templatate, sense, coding
Biology 1A03 Cellular and Molecular Biology
NOTES
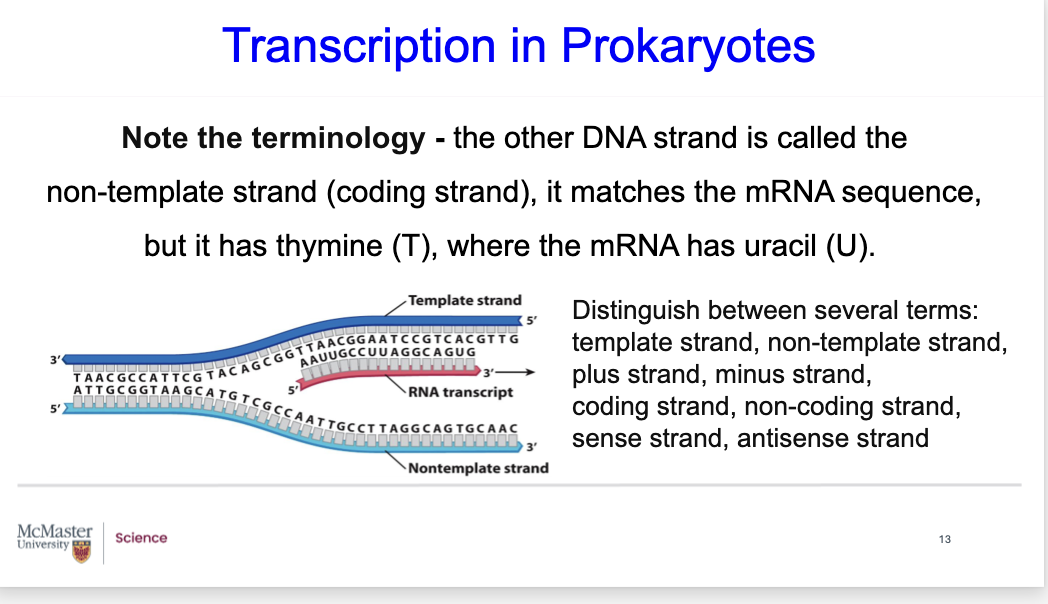
Transcription in Prokaryotes
Note the Terminology - since the other DNA strand is called the non-template strand (also known as the coding strand),
it matches the mRNA sequence but it has thymine (T), where the mRNA has uracil (U).
Please note that there are several different terms that you should know:
- the template strand and the non-template strand
- the plus strand and the minus strand
- the coding strand and the non-coding strand
- the sense strand and the antisense strand.
Make sure that you are able to distinguish between each of these.
Prokaryotic RNA Polymerase: What is the structure of RNA Polymerase ?
Researchers used X-ray crystallographic techniques to study the 3Dstructure of RNA polymerase
It is a globular protein with channels running through it.
The active site, where phosphodiester bonds are formed, is found atthe intersection of several channels.
Researchers used X-ray crystallographic techniques to study the 3D structure of RNA polymerase
It’s a globular protein, which has channels running through it.
The active site, where phosphodiester bonds are formed, is found at the intersection of several channels.
what is Holoenzyme?
RNA Polymerase (Core Enzyme) + Sigma (protein)
= Holoenzyme
In prokaryotes, the RNA Polymerase (Core Enzyme) associates with Sigma (protein), which collectively forms the holoenzyme. EUKARYOTES DON’T HAVE SIGNMA
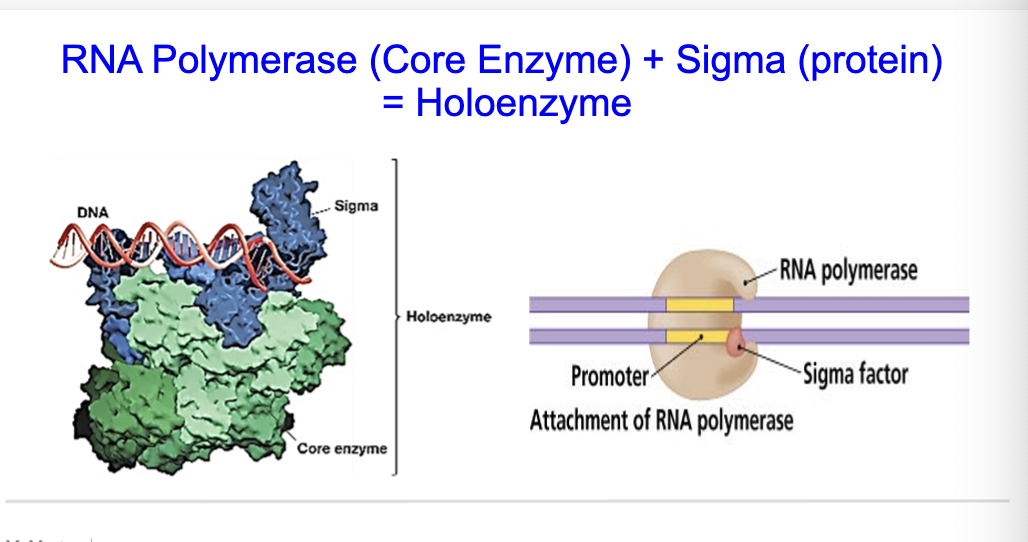
How does Transcription begin in bacteria?
An E.coli protein called Sigma binds to RNA polymerase beforetranscription begins.
RNA Polymerase + Sigma = Holoenzyme
Holoenzyme binds to DNA at specific sites called promoters.
Prokaryotic RNA Polymerase
How does Transcription begin in bacteria?
An E.coli protein, Sigma, binds to RNA polymerase before transcription begins.
RNA polymerase is the core enzyme
RNA Polymerase + Sigma = Holoenzyme
Holoenzyme binds to DNA at specific sites called promoters.
What is the function of Sigma ?
Sigma is required to facilitate RNA polymerase binding to DNA
Sigma helps to guide RNA Polymerase to promoters
Sigma is required to facilitate RNA polymerase binding to DNA.
Sigma helps to guide RNA Polymerase to promoters, even before transcription starts.
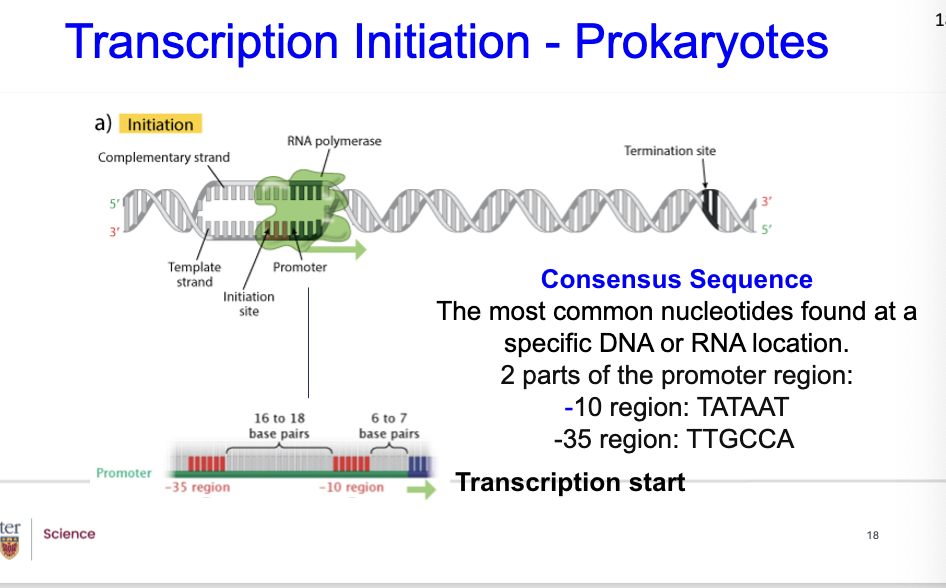
What are Promoters?
They are sites on the DNA template, where transcription begins.
what is consensus sequences?
The most common nucleotides found at aspecific DNA or RNA location.
2 parts of the promoter region:
-10 region: TATAAT
-35 region: TTGCCA
Consensus Sequences - The most common nucleotides found at a specific DNA or RNA location.
what is the analogy to understand downstream and upstream terminology?
Analogy to understand downstream and upstream terminology.
Imagine that you are standing on a river’s side bank and you throw a ball in the water and it flows with the current downstream. If it is downstream , it is downstream from that start position. Whereas, if it is upstream, it is located before your start position on a riverbank.
What is the structure of Promoters?
Pribnow analyzed the base sequences of promoters.
Found that promoters have the sequence TATAAT in common.This sequence is located 10 bases upstream from where RNApolymerase starts transcription, it’s the -10 region.
Another conserved sequence, TTGCCA, occurs in the samepromoters 35 bases upstream of the start site, it’s the -35 region.
Transcription Initiation - Prokaryotes: How many types of Sigma proteins do bacteria have?
Bacteria have several different types of Sigma proteins, eachbinds to different promoters, determining which genes aretranscribed during a given time.
Transcription Initiation - Prokaryotes: What happens when Sigma binds to the promoter?
DNA double helix opens, the template strand is threaded through thechannel in RNA Polymerase that leads to the active site.
Ribonucleotide triphosphates enter another channel in RNA Polymerase, move to the active site, & are incorporated into the mRNAwhen they are complementary to the template strand.
what happens in
Elongation of Transcription - Prokaryotes
Elongation of Transcription – Prokaryotes
RNA Polymerase moves 3’ to 5’ along the template DNA strand:
It unwinds the double helix
It reads the DNA template and brings in the complementary nucleotide.
It catalyzes the addition of the nucleotides to the 3’end of the growing RNA molecule.
It inserts an uracil when it encounters adenine in the template DNA, since in mRNA there is uracil, instead of thymine.
what is happening in this diagram?
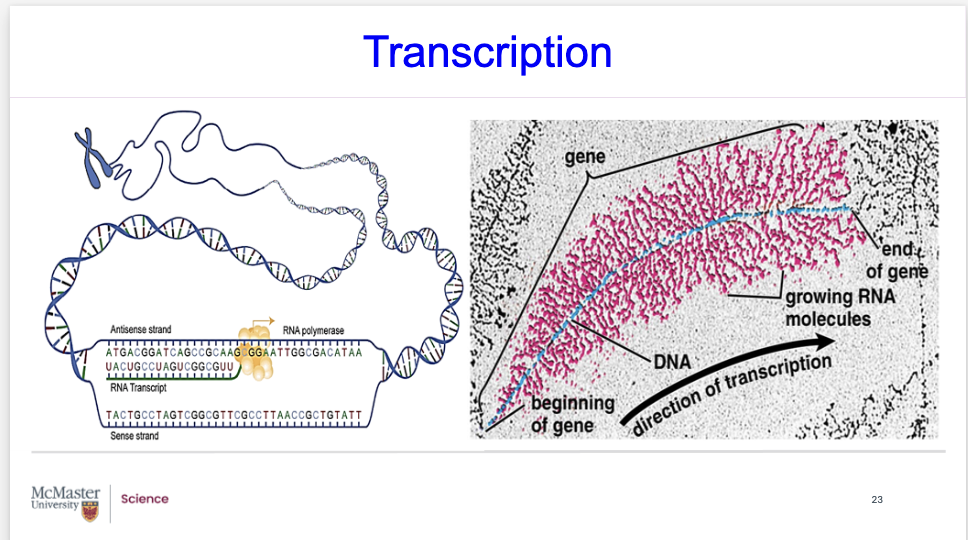
Biology 1A03 Cellular and Molecular Biology
NOTES
Transcription
Looking at the top figure, you can see that you need to distinguish the terminology.
You should know what is the antisense strand and what is the sense strand.
Here’s another diagram, that we can see the direction of transcription.
A wonderful TEM that shows the direction of transcription and the growing RNA molecules.
what happens in the
Termination of Transcription - Prokaryotes?
Specific sequences in template DNA act as termination sites.Bases in the transcriptional termination sequence formcomplementary base pairs.
This results in hairpin structure, which causes the RNAstrand to separate from the RNA Polymerase, terminatingtranscription. RNA polymerase & mRNA are released.
Specific termination proteins may facilitate the release insome bacteria.
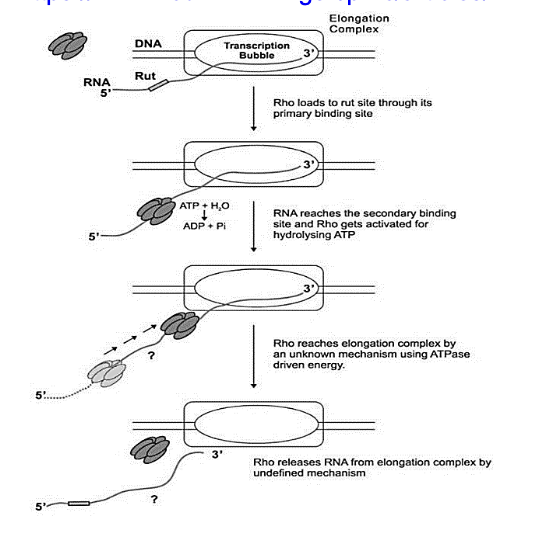
What happens in rho dependent termination of transcription?
2. Rho-dependent termination uses a termination factor known as a Rho protein which binds to the site of termination
and disassembles the mRNA from the DNA as well as the transcription machinery. Rho is a helicase enzyme which
depends on energy from ATP through the use of ATPase activity to drive this disassembly. The article below describes
how the Rho enzyme facilitates this but I will provide a very brief summary. Rho seems to have an affinity for pyrimidines
over purines, especially cytosine. With that being said, there is a rut site on the mRNA which is cytosine rich and non-
translating to allow for the binding of Rho. Through this, Rho travels along the transcript via undefined mechanisms to
facilitate the disassembly of the RNA-DNA duplex. Here is a supplemental image as well as the research article. Feel
free to delve deeper as this article highlights quite a lot of interesting components to Rho-dependent termination.
However, please note that these mechanisms are still being researcher
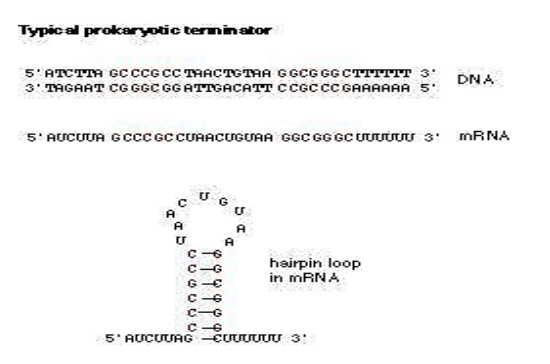
What happens in rho independent termination of transcription?
1. Rho-independent termination in prokaryotes uses a termination sequence which contains inverted repeats on the
DNA template strand. This would include repeats of C and G nucleotides to then be reciprocated onto the growing mRNA
strand. With that, the mRNA forms a GC rich hairpin loop where it forms base pairs with itself and allows for the
disassembling of the transcription machinery as well as the mRNA from the DNA. Below is a supplemental image which
portrays the GC rich hairpin loop as well as the termination sequence with inverted repeats to display the origin of the
loop
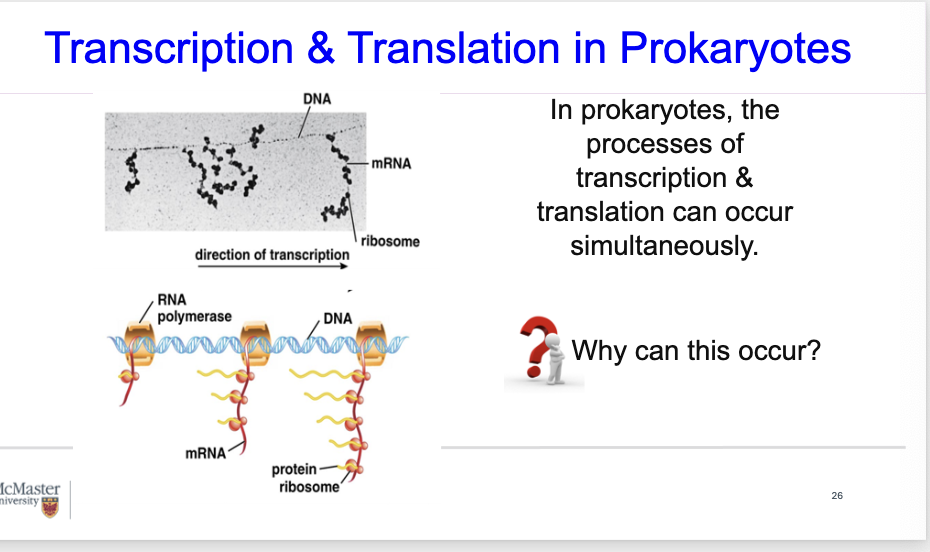
In prokaryotes, theprocesses of
transcription &translation can occursimultaneously.
Why can this occur?
Transcription & Translation in Prokaryotes
In prokaryotes, transcription & translation occurs simultaneously.
TEM is shown at the top.
In the figure at the bottom, you see a number of transcripts shown in red, and shown in yellow are the polypeptides that are produced. One on the left is more recent , compared to the one on the right which is more progressed.
Why can this occur?
In prokaryotes, you recall that transcription and translation are not separated in time and space, so that they can occur simultaneously. It can occur very fast, and it does not have to go out of the nuclear envelope like what occurs in eukaryotes.
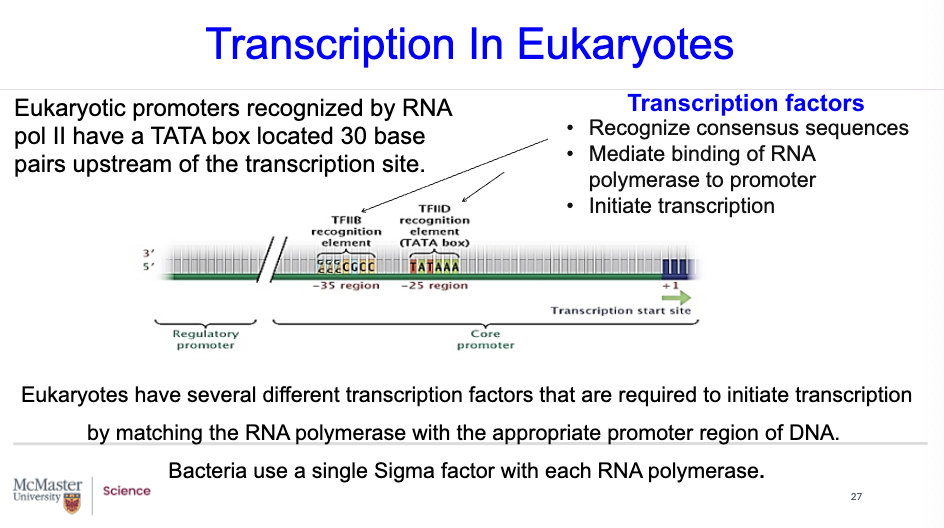
In eukaryotes, when they are going through transcription, do they have Sigma?
No, eukaryotes don’t have sigma. Sigma is mentioned when we are considering prokaryotes.
In terms of eukaryotes, we say that they have transcription factors.
Transcription factors
Recognize consensus sequences
Mediate (regulate & control) binding of RNA polymerase to promoter
Initiate transcription
Eukaryotes have several different transcription factors that are required to initiate transcription by matching the RNA polymerase with the appropriate promoter region of DNA.
In comparison, prokaryotes like bacteria use a single Sigma factor with each RNA polymerase
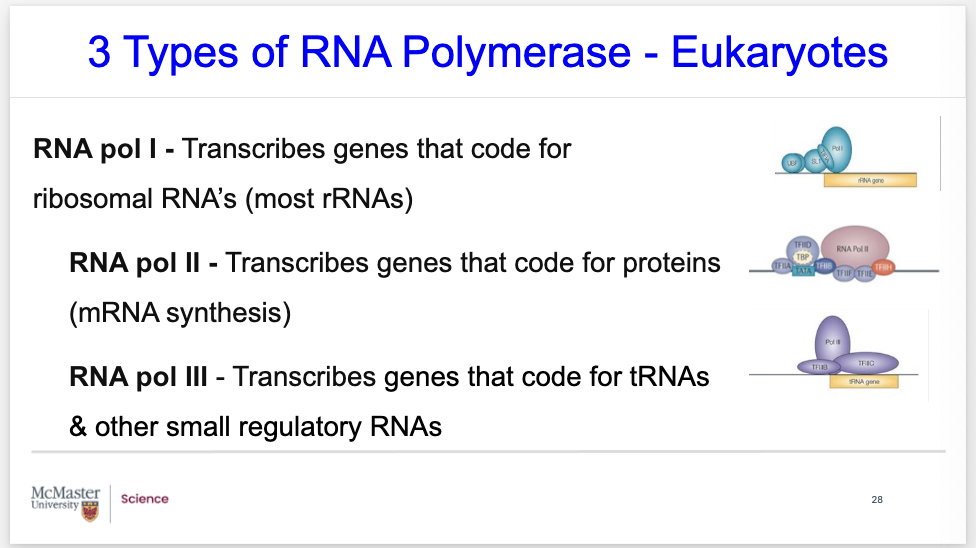
what are the 3 types of RNA polymerase- eukaroyotes?
Biology 1A03 Cellular and Molecular Biology
NOTES
3 Types of RNA Polymerase - Eukaryotes
RNA pol I - Transcribes genes that code for ribosomal RNA’s (most rRNAs)
RNA pol II - Transcribes genes that code for proteins associated with mRNA synthesis. We are very much interested in this one with regards to the process of transcription.
RNA pol III - Transcribes genes that code for transfer RNAs (tRNAs) & other small regulatory RNAs. We will be interested in reviewing this RNA polymerase III with regards to translation.
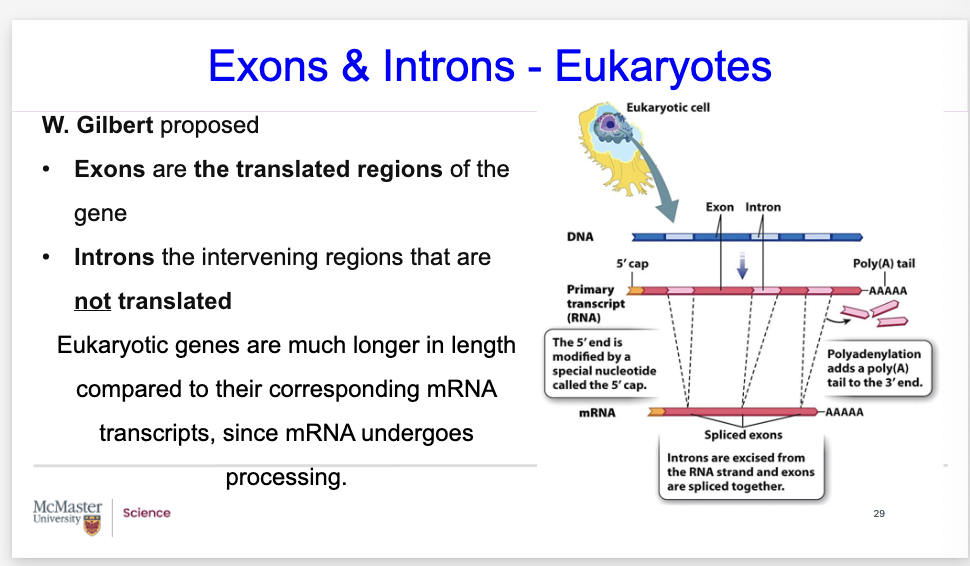
What are exons and introns? What is W gildbert propose?
How are Eukaryotes also different from Prokaryotes?
In eukaryotes, you have RNA processing that occurs.
Introns & Exons - Eukaryotes
W. Gilbert proposed that the expressed portions of genes = exons & the intervening portions = introns.
Make sure that you understand these two types:
Exons are translated regions of the gene
Introns are not translated regions of the gene
Eukaryotic genes (DNA) are much longer in length compared to their corresponding mRNA transcripts, since mRNA undergoes processing.
The introns are spliced out of the primary transcript RNA and, the exons that are spliced together, and later are translated to form the polypeptides.
Post-Transcriptional Processing of Eukaryotic mRNAs?
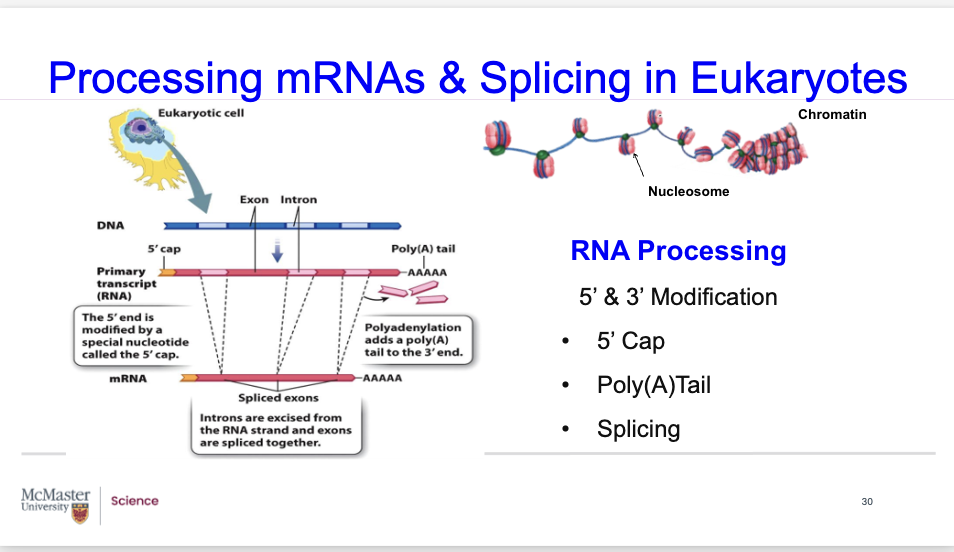
Processing mRNAs & Splicing in Eukaryotes
RNA Processing involves the following:
5’ & 3’ Modification
The additions of 5’ Cap & a Poly(A)Tail.
The removal of the introns and splicing together of the exons.
Post-Transcriptional Processing of Eukaryotic mRNAs
5’ Cap
a 7-methylguanosine is attached to the mRNA through an unusual 5’ to 5’triphosphate linkage
functions as a recognition signal for translation machinery & protects thetranscript from degradation
3’ Poly (A) tail
Consists of 150-200 adenines is added to the 3’ end of the mRNA
functions to protect the transcript from degradation.
What is intron splicing?
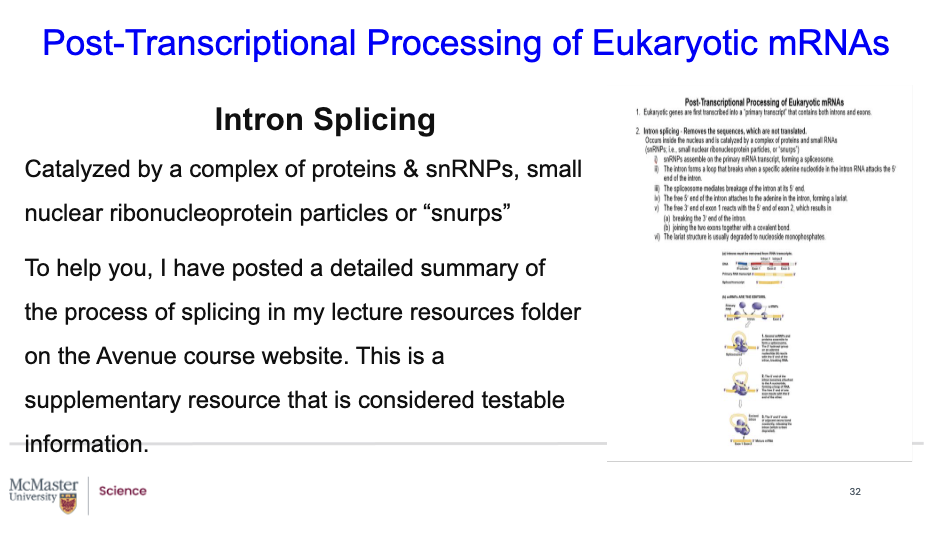
Biology 1A03 Cellular and Molecular Biology
NOTES
Post-Transcriptional Processing of Eukaryotic mRNAs
Intron Splicing
Catalyzed by a complex of proteins & snRNPs, small nuclear ribonucleoprotein particles or “snurps”
To help you I have posted a detailed summary of the process of splicing in my lecture resources folder on the Avenue course website. This is a supplementary resource that is considered testable information
what evidence led ot the conclusion of triplet nucleiotides? who discovered this?
Crick, Barnett, Brenner, & Watts-Tobin provided conclusive evidence that the genetic code is written out in triplets of nucleotides. If a single nucleotide was inserted within the nucleotide sequence, the code was misread and the series of codons after that added nucleotide were different from the original series. Also, a similar effect would happen if one nucleotide was removed. And the researchers observed the same effect when 2 nucleotides were added or removed.
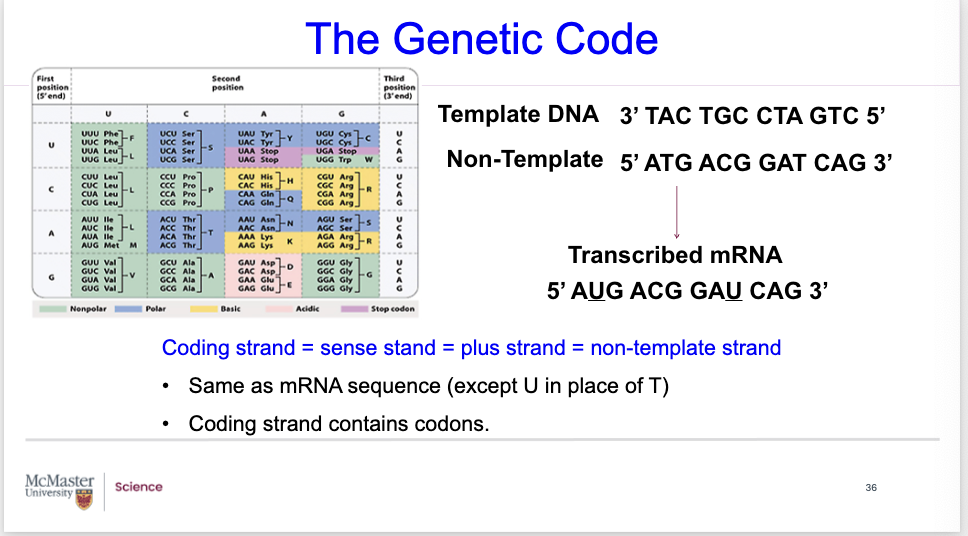
proper terminology of template and non template strands
Biology 1A03 Cellular and Molecular Biology
NOTES
The Genetic Code
You should be able to distinguish the:
Template DNA strand
Non-Template strand
Transcribed into mRNA
You need to know the proper terminology.
Coding strand is also known as the sense stand, and the plus strand, and the Non-template strand.
Note that the coding strand (non-template) has the same sequence as the mRNA sequence (except for the DNA there is T, whereas in mRNA there is U.
Coding strand contains codons.
what is ORF?
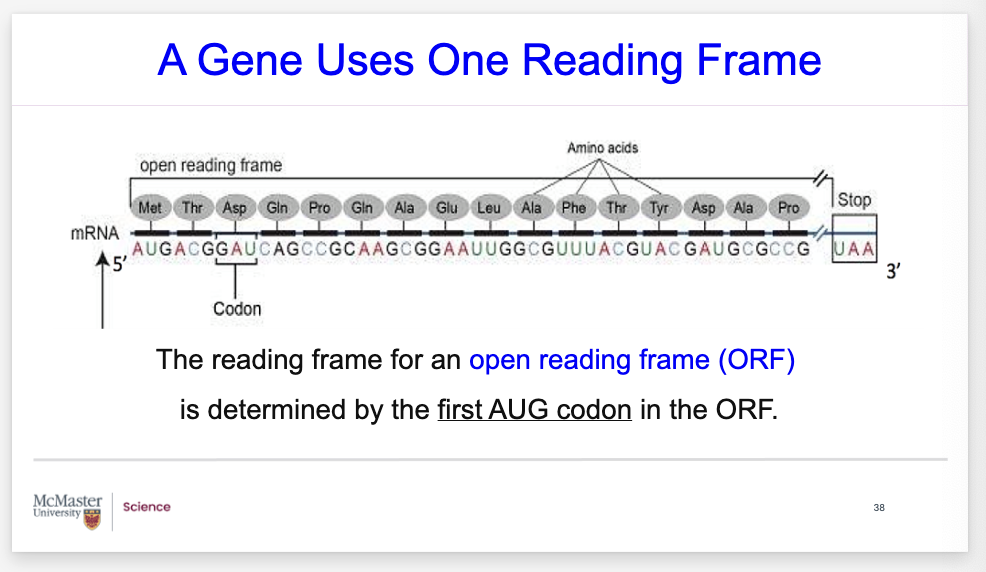
Biology 1A03 Cellular and Molecular Biology
NOTES
A Gene Uses One Reading Frame
In this figure, the reading frame for an open reading frame (ORF) is determined by the first AUG codon in the ORF.
This gives a point of reference and location for the gene of interest.
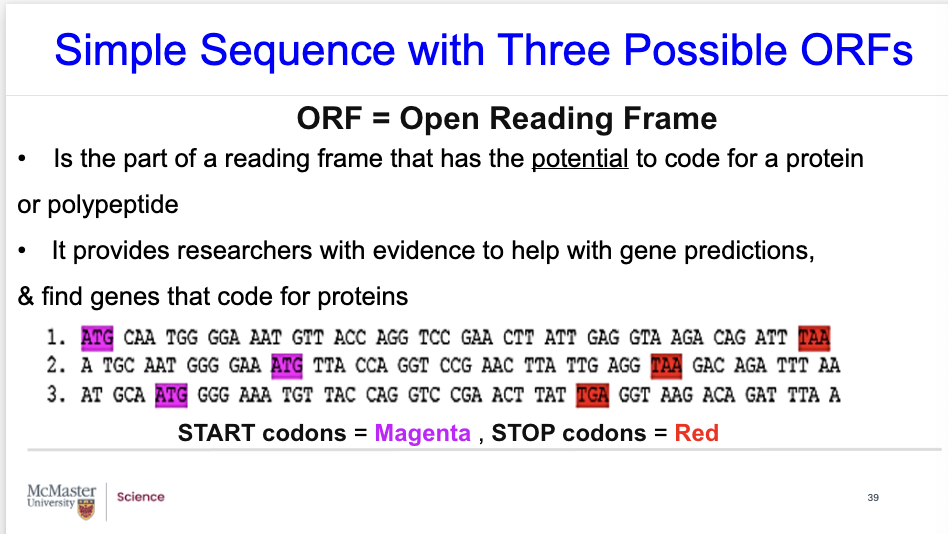
Open reading frame is abbreviated ORF.
Is the part of a reading frame that has the potential to code for a protein
It provides researchers with evidence to help with gene predictions, and find genes that code for polypeptides.