FMLec | M2 Culture-Dependent Techniques, APC & EAPC Calculations
1/75
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
76 Terms
2 general ways to detect organisms in food
Culture-dependent techniques
Non-culture-dependent techniques
11 culture-dependent techniques
ct3m adrd mm
Conventional standard plate counts (SPC/APC)
Turbidimetric measurement
Most probable number (MPN)
Membrane filtration
Microscope colony count
Agar droplet count
Dye reduction
Roll tubes
Dry film and related methods
Microbiological examination of surfaces
Metabolically injured microorganisms
Culture-dependent technique
_ is also known as aerobic plate count (APC)
Used to count the number of viable cells (i.e., those capable of forming an offspring) and thus the number of colony forming units (CFU) because the assumption is each colony arises from 1 viable cell
Conventional standard plate count (SPC)
Why do we count CFUs in SPCs?
It’s because of the assumption that each viable cell can grow and divide to yield one colony, 1 cell = 1 colony
When is APC or SPC used?
For counting the no. of viable cells = no. of CFUs since 1 cell = 1 colony
7 factors affecting viable count (in A/SPC)
sdn ppti
Sampling methods employed
Distribution of organisms in the food sample
Nature of food biota and food material
Pre-examination history of the food product
Allows us to know the (1) relative number of organisms in food sample, (2) existence of other competing or antagonistic organisms
Plating medium used
Nutritional adequacy, pH, oxidation-reduction potential (Eh) of the plating medium, water activity npow
Type of diluent
Incubation time and temperature used
Explain SPC procedure
Food sample homogenization
Serial dilution
Plating in or onto a suitable agar medium
Incubation at appropriate temperature (for specific period of time)
Counting of colonies
Calculation of CFU/g or CFU/mL
2 equipments which may be used for food sample homogenization
Stomacher
Mechanical blender
_ homogenizes specimen in a special plastic bag
Vigorous pounding of 2 paddles that shear the food specimen
Microorganisms are then released into the diluent
Stomacher
4 advantages of stomacher over mechanical blender
mens
More pleasant noise level
Easy to clean
No heat buildup
Allows for the storage of homogenates in the freezer
_ is a stepwise process of reducing the concentration of microorganisms in a sample
Serial dilution
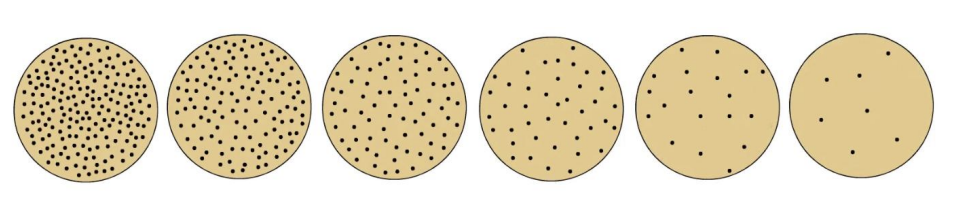
Explain the principle behind serial dilution and the precautionary measure prior to doing it
Serial dilution will thin out microbial population until there is only 1 cell left in a tube of diluent (10-fold decrease in no. of cells as dilution increases)
Prior to transferring inoculum from 1 tube to another, it’s important to vigorously shake the tube where you’ll be taking your inoculum from, specifically shaking it 25 times in 1 ft arc within 7s or use a vortex mixer
3 major sources of breast milk microbiome
Child’s oral cavity
Presence of oral bacteria in breast milk (e.g., Streptococcus salivarius, S. mitis, Gemella, Rothia)
Reverse flow of milk from infant’s mouth → woman’s milk ducts
Mother’s skin
Presence of commensal bacteria (e.g., S. epidermidis, Corynebacterium, Malassezia)
Colonization of mammary gland with mother’s skin microbiota thru nipple
Mother’s digestive tract
Presence of anaerobic bacteria of GI tract (e.g., Bifidobacteria, Bacteroides, Clostridium, Saccharomyces)
Entero-mammary route during pregnancy and lactation
In plating step (in or onto a suitable agar medium), 2 factors must be considered including _
Type of media (based on composition and use)
Plating technique
Types of media based on composition
Defined media
Exact quantitative and qualitative composition
Prepare by adding precise amounts of highly purified organic or inorganic chemicals to distilled water
Simple = if it contains only 1 carbon (C) source
Complex = if more than 1 C source
Complex media
Contain digests of microbial, animal, or plant products, or any number of other highly nutritious yet impure substances
e.g.,
Casein (milk)
Beef extract
Peptone (protein hydrolysate)
Tryptic soy broth (soybeans)
Yeast extract
Explain the types of media based on use/purpose
General purpose media (e.g., Nutrient Agar)
Supports almost all microbial growth
Do not contain inhibitory substances
Enriched media (e.g., Blood Agar)
Growth stimulants, e.g., blood, serum, other highly nutritious substances bso
Increases the number of desired microorganisms to a detectable level but does not suppress the growth of other bacteria
For growing fastidious microorganisms (i.e., those with complex nutritional requirements)
Selective media (e.g., Mannitol Salt Agar)
Inhibits growth of unwanted microorganisms and encourages growth of a particular organism
Contains inhibitors, e.g., antibiotics, dyes, toxic compounds, detergents adtd
Differential media (e.g., Eosin Methylene Blue Agar)
Distinguishes different species of bacteria, usually containing indicators (e.g., dyes, pH indicators) that reveal differences in microbial metabolism and enzymatic activity
Type of media based on purpose/use
Contains growth stimulants, e.g., blood, serum, other highly nutritious substances
For growing fastidious microorganisms
Increases the number of desired microorganism to a detectable level but does not suppress the growth of other bacteria
Enriched media
Type of media based on purpose/use
Distinguishes different species of bacteria
Usually contains indicators (e.g., dyes) that reveal differences in microbial metabolism and enzymatic activity
Differential media
Type of media based on purpose/use
Supports almost all microbial growth
Does not contain any inhibitory substances
General purposed media
Type of media based on purpose/use
Contains inhibitory substances (e.g., antibiotics, dyes, toxic compounds, detergents)
Inhibits the growth of unwanted microorganisms while encouraging the growth of a particular organism
Selective media
T/F: Enriched media contains inhibitory substances
FALSE
While enriched media contains growth stimulants, it only serves to increase the number of desired microorganism to a detectable level but does not suppress growth of other bacteria
Type of media based on purpose/use
e.g., Eosin Methylene Blue Agar (EMBA)
Differential media
Type of media based on purpose/use
e.g., Nutrient Agar
General purpose media
Type of media based on purpose/use
e.g., Mannitol Salt Agar
Selective media
Type of media based on purpose/use
e.g., Blood Agar
Enriched media

Defined or Complex media?
Complex media (with yeast extract, peptone)

Defined or Complex media?
Simple defined media (only 1 C source)
Type of media (based on composition) with exact quantitative and qualitative composition, prepared by adding precise amounts of highly purified organic or inorganic chemicals to dH2O
Defined media
Defined media with only 1 C source
Simple defined media
Defined media with more than 1 C source
Complex defined media
3 plating techniques
Spread plating (0.1 mL or 100 uL inoculum)
Pour plating (1 mL or 1000 uL)
Miles and Misra / Drop Count technique (0.01 mL or 10 uL)
Explain steps to spread plating
Pipette 0.1 mL (100 uL) sample into solidified agar surface of plate
Spread sample evenly over surface using L rod and turn table
Incubate
Colonies grow on surface
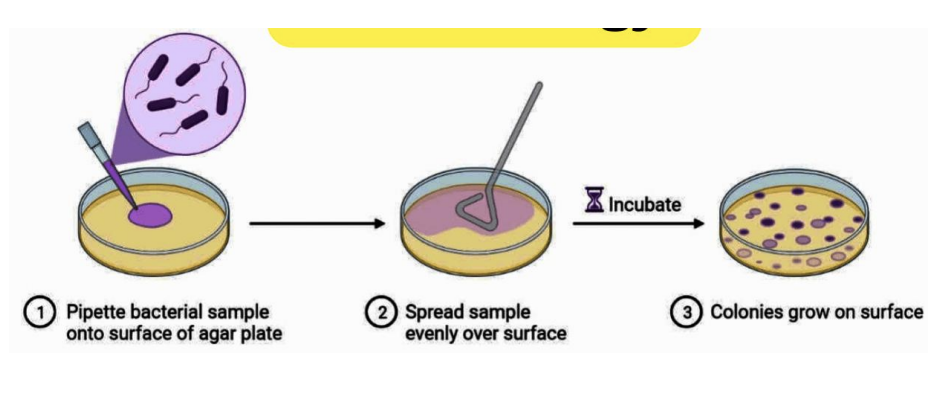
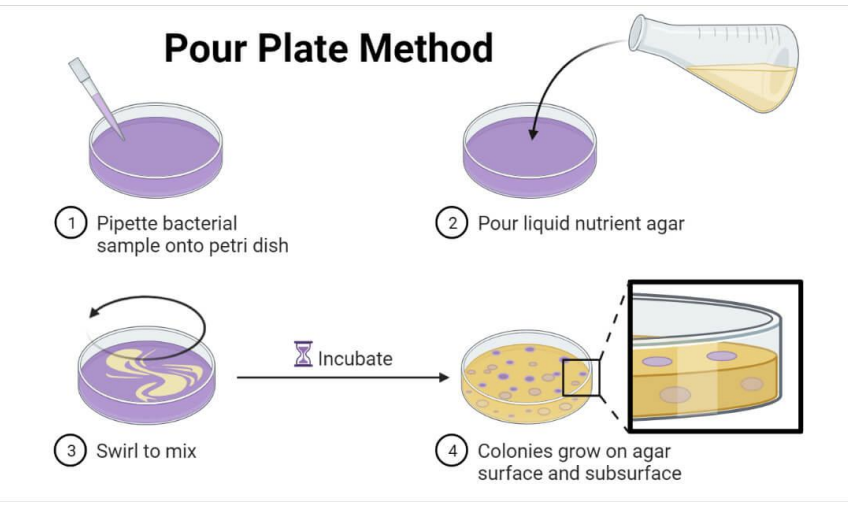
Explain steps to pour plating
Pipette 1 mL (1000 uL) sample into plate
Pour molten / liquefied agar
Swirl to mix (do figure-8 pattern)
Incubate
Colonies grow on surface and subsurface
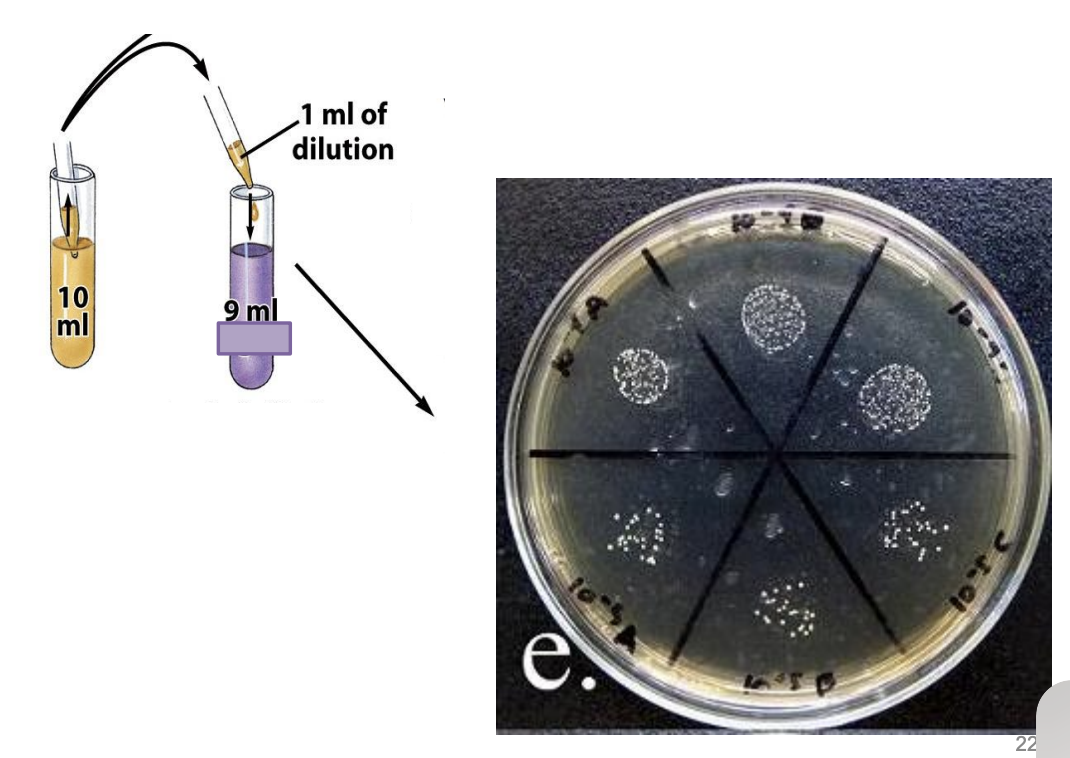
Explain steps to Miles and Misra / Drop Count technique
Serially dilute sample
Divide plate into 6 parts (or into how many dilutions you’ll plate)
Pipette 0.01 mL (10 uL) of sample of particular dilution into each division of plate
Incubate
Colonies grow on surface
Spread vs. Pour vs. Miles and Misra / Drop Count Technique
cbd tvma | Spread plate | Pour plate | Miles and Misra |
Culture media | Pre-solidified media | Molten/liquefied media | Pre-solidified agar |
Basis of isolation | Serial dilution, Spatial separation | Serial dilution, Spatial separation | Serial dilution |
Desired microorganisms | Present at higher level than any other microorganism | Present at higher level than any other microorganism | Present at higher level than any other microorganism |
Type of colonies | Surface | Surface/subsurface | Surface |
Vol of inoculum | 0.1 mL (100 uL) | 1 mL (1000 uL) | 0.01 mL (10 uL) |
Main lab tool | L rod/Bent glass rod/hockey sticks, Micropipette, Turntable | Micropipette, | Micropipette |
Application | Isolation, Enumeration (Heat-sensitive aerobes) | Isolation, Enumeration (Heat-tolerant aerobes, Microaerophiles) | Enumeration (Heat-sensitive aerobes) |
_ refer to the 3 key temperature points that define the growth range of a microorganism
Cardinal temperatures
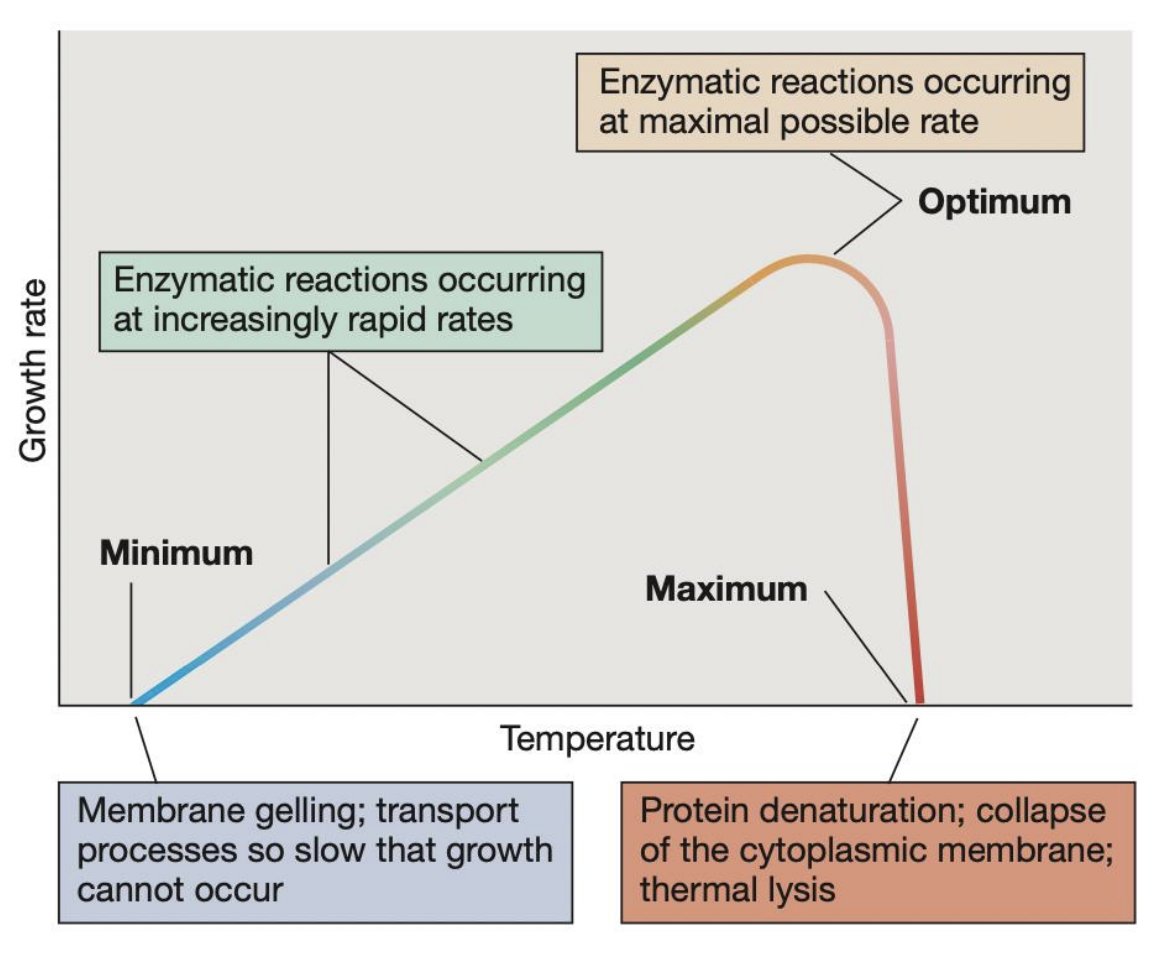
_ refers to the highest temperature at which microbial growth can occur; beyond this, protein denaturation, collapse of cytoplasmic membrane, and thermal lysis may occur pct
Maximum temperature
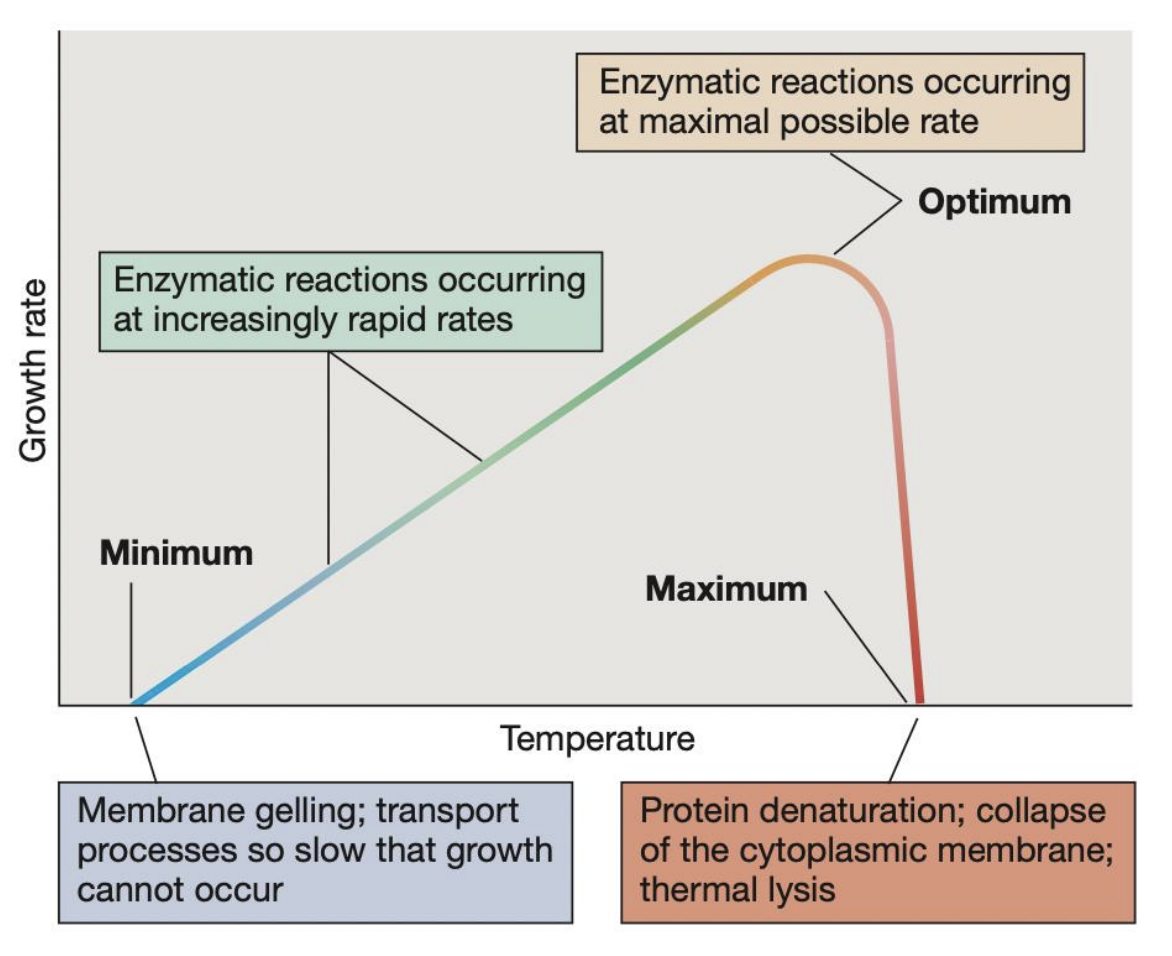
_ refers to the temperature at which growth rate is fastest and enzymatic reactions occur at maximal possible rate
Optimum temperature
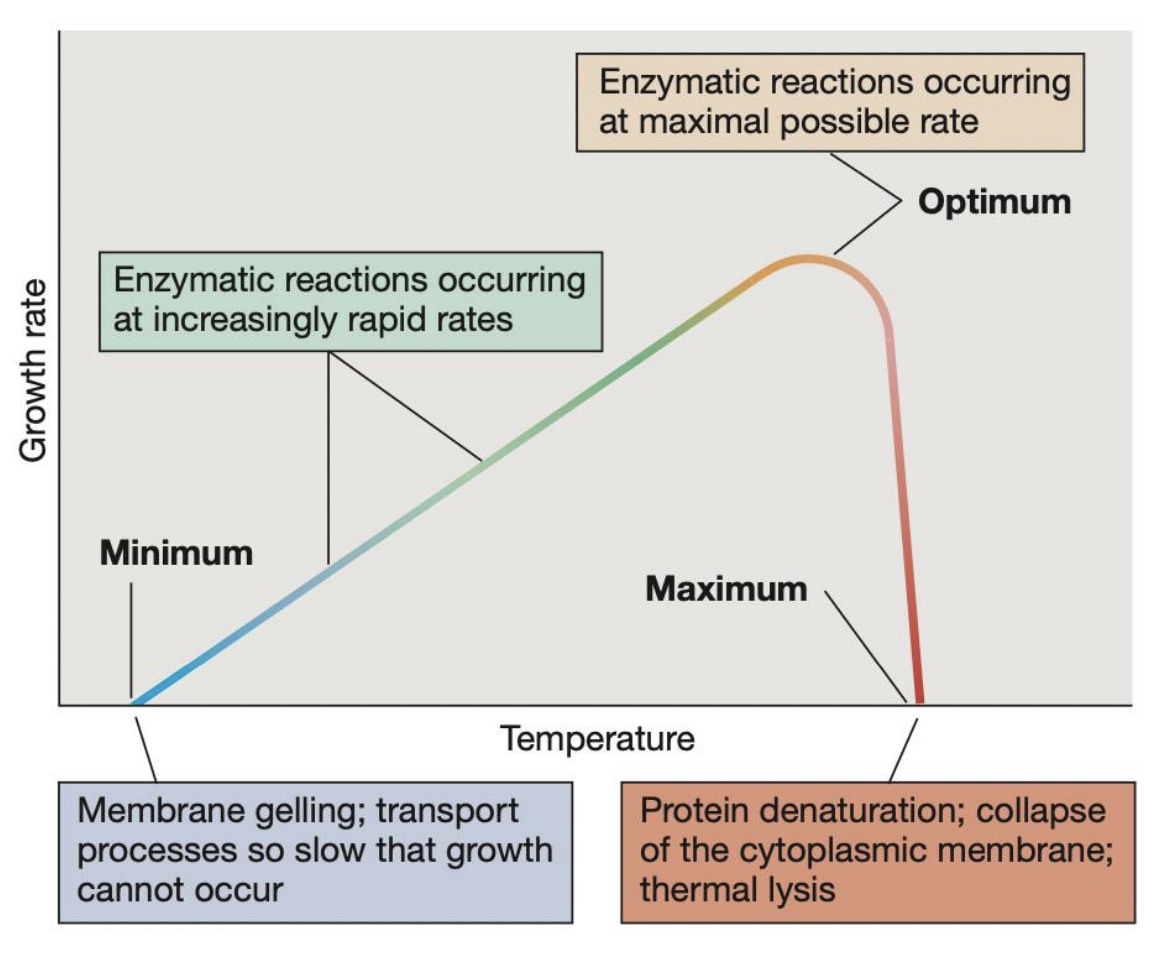
_ refers to the lowest temperature at which microbial growth can occur; beyond this, membrane gelling and transport processes become so slow that growth cannot occur
Minimum temperature
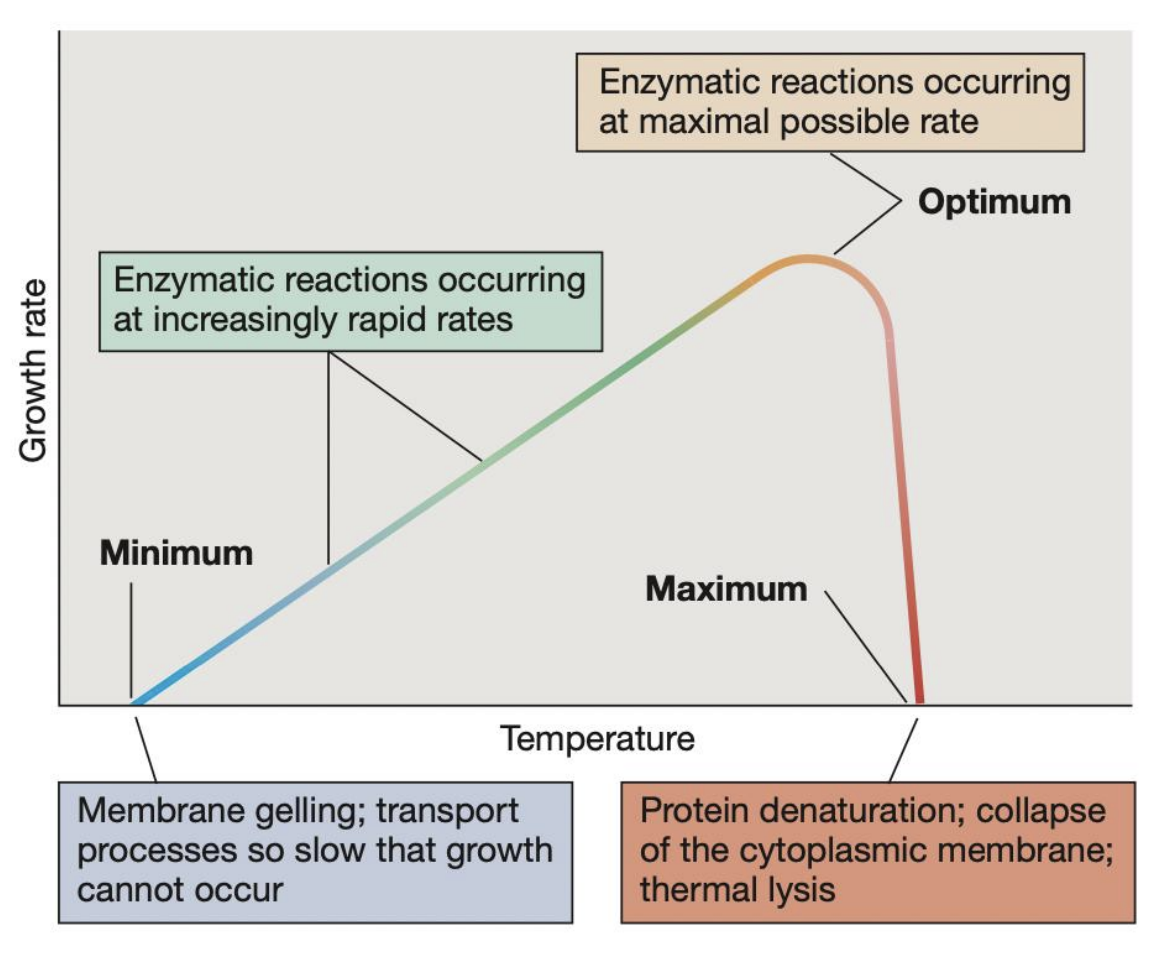
Different types of organisms based on their preferred temperature range
Psychrophiles = 4 C Polaromonas vacuolata
Psychrotolerant
Mesophiles = 39 C E. coli
Thermophiles = 60 C Geobacillus stearothermophilus
Thermotolerant
Hyperthermophiles = 88 C Thermococcus celer, 106 C Pyrolobus fumarii
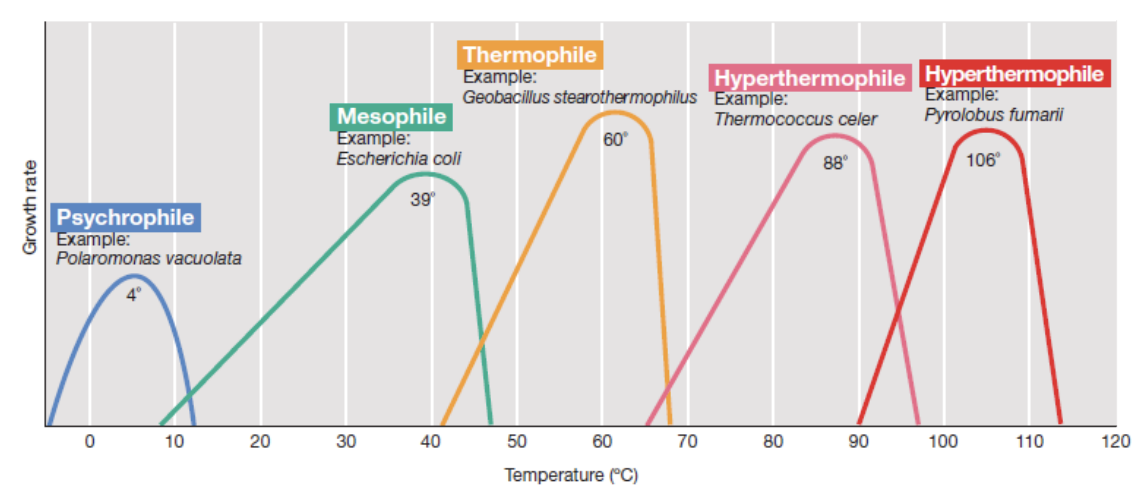
A microbe incubated beyond its maximum (cardinal) temperature may experience _
pct
Protein denaturation
Collapse of the cytoplasmic membrane
Thermal lysis
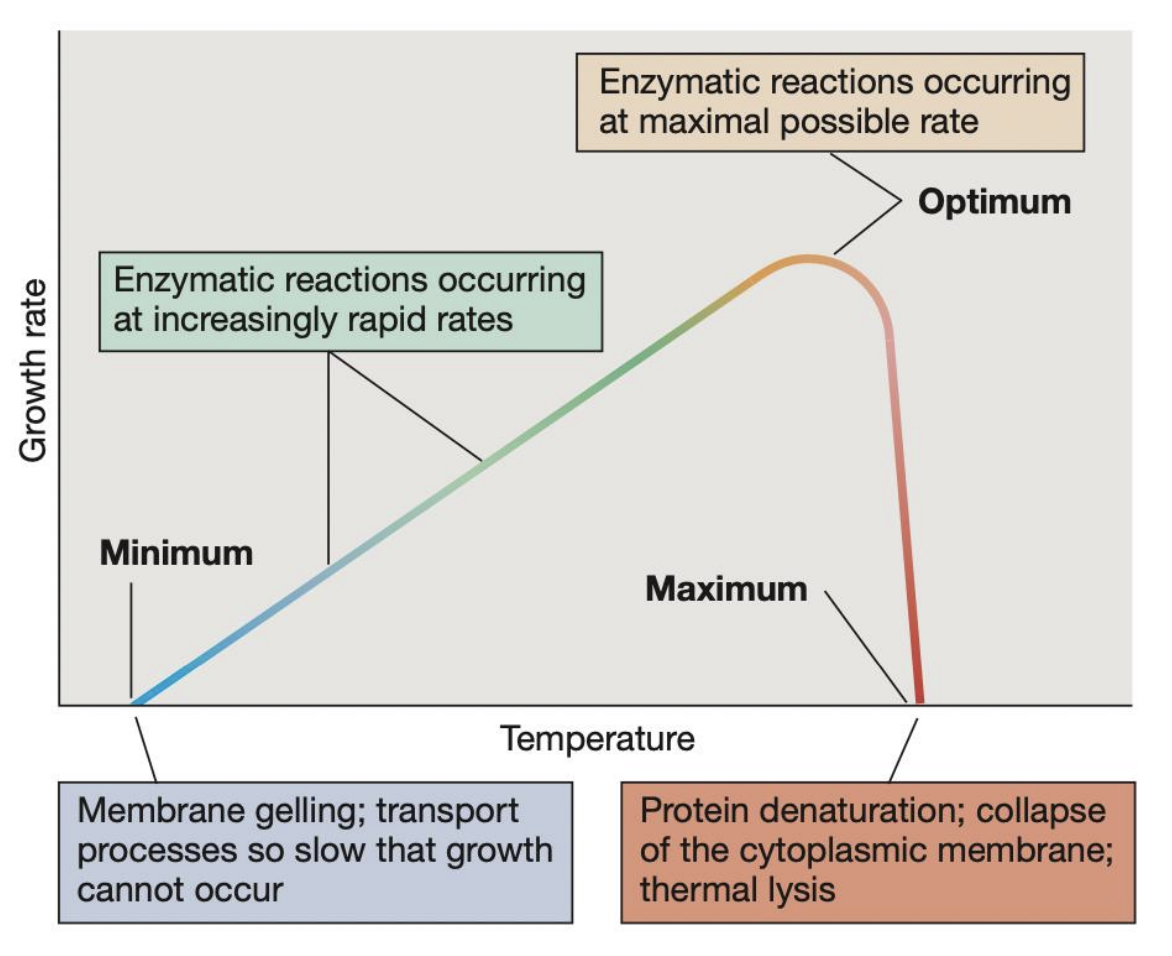
A microbe incubated at lower its minimum (cardinal) temperature may experience _
membrane gelling and transport processes may be so slow that growth cannot occur
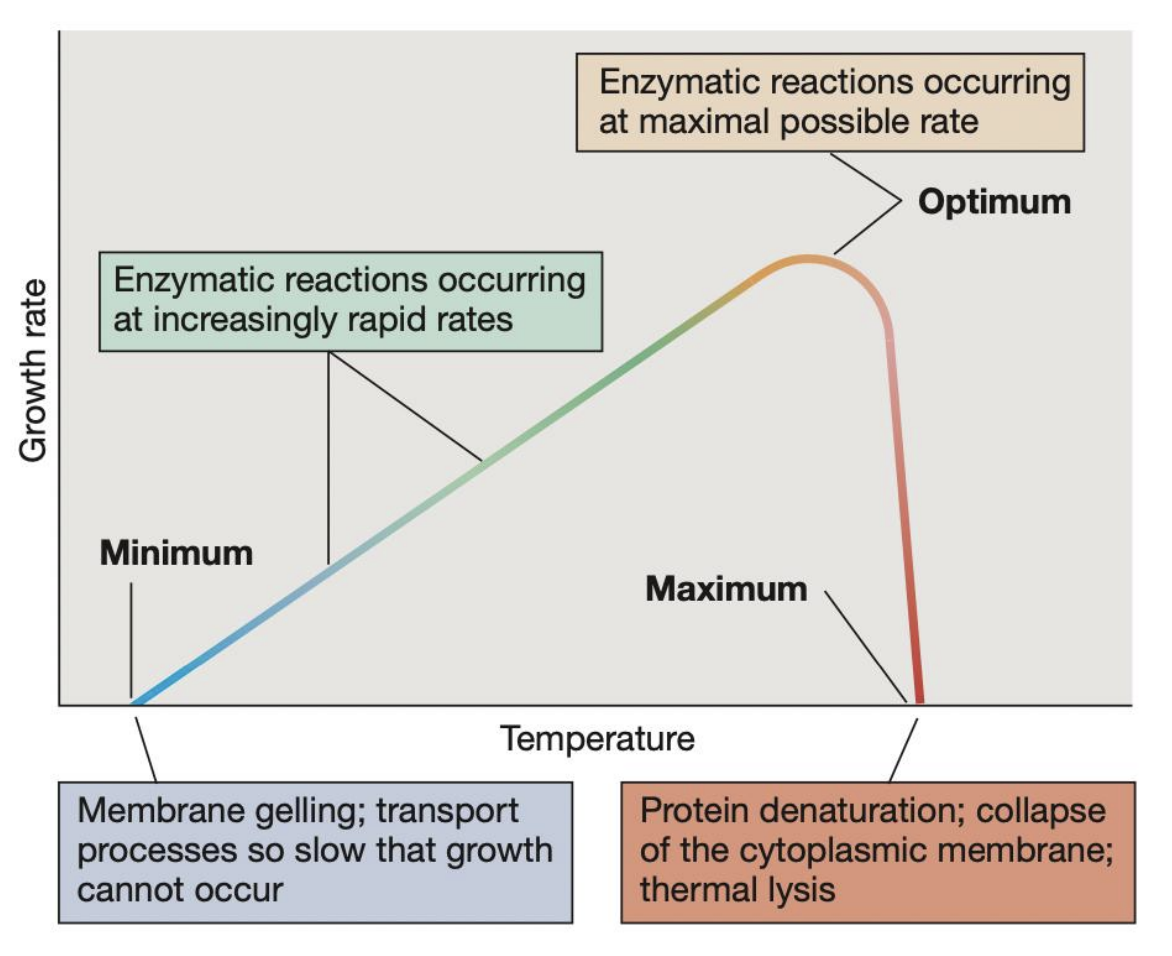
In a microbe incubated at its optimum temperature, _ may be observed
enzymatic reactions occurring at maximal rate
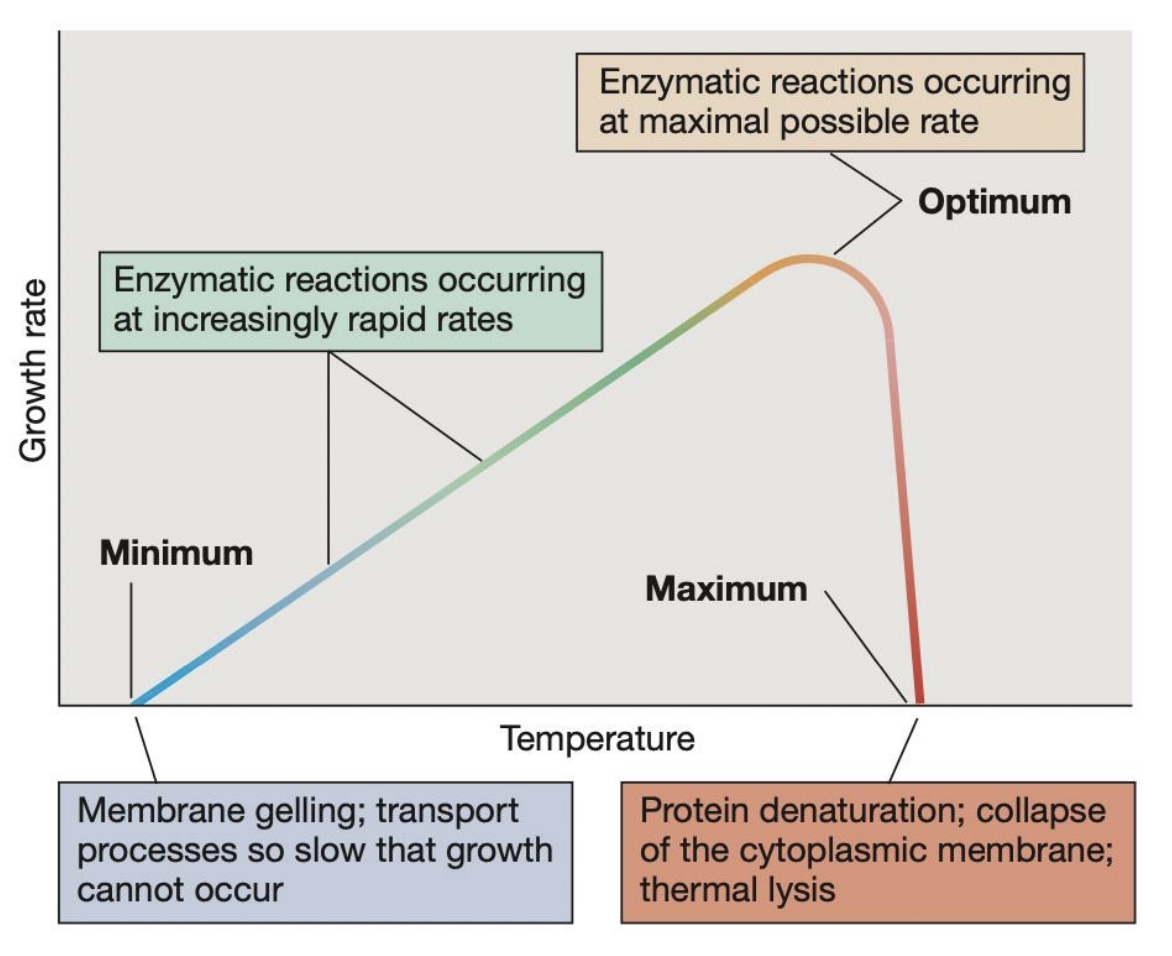
T/F: Mesophiles can grow at 0°C but will have significantly slower growth rates
FALSE
Mesophiles generally cannot grow at 0°C. Psychrotrophs and psychrophiles, however, can.
T/F: Psychrophiles and psychrotrophs can both grow at low temperatures, but only psychrotrophs can tolerate higher temperatures beyond 20°C
TRUE
Psychrotrophs have a broader temperature range and can tolerate higher temperatures, unlike psychrophiles, which die at warmer temperatures.
Explain 5 different types of microorganisms based on oxygen requirements
Obligate aerobes (Micrococcus luteus)
Need O2 to survive
Facultative anaerobes (E. coli)
Can survive without O2 but prefers it when available
Aerotolerant (Streptococcus mutans)
Do not use O2 but can tolerate it
Strict anaerobes (Methanobacterium formicicum)
Exposure to O2 is toxic
Microaerophiles (Spirillum volutans)
Require O2 at low concentrations but atmospheric O2 is harmful to them
T/F: A facultative aerobe and a facultative anaerobe refer to two different types of organisms
FALSE
These terms are used interchangeably; they both refer to organisms that can survive without O2 but prefers it
T/F: Aerotolerant anaerobes and facultative anaerobes both grow in oxygen, but only facultative anaerobes use it for metabolism
TRUE
Facultative anaerobes use aerobic respiration when oxygen is available, while aerotolerant anaerobes never use oxygen but tolerate it.
T/F: All microaerophiles can survive at normal atmospheric oxygen levels (21%)
FALSE
They require low oxygen levels (~2-10%) and are harmed by high oxygen concentrations.
Where to incubate
Aerobes, facultative anaerobes, aerotolerant
Normal incubator
Where to incubate
Microaerophiles
Microaerophilic jar or can (has candle inside to keep O2 levels low)
Where to incubate
Strict anaerobes
Anoxic jar
Anoxic glove box
Make-shift jar with oxygen absorber inside
Methods that may be used for colony counting
Colony counter equipment
Mobile applications
Manually using pen
3 important guidelines in colony counting
Count all colony forming units (CFUs), including pinpoint
Count immediately after incubation period
Store plates at 4C for not more than 24 hrs if it’s impossible to count at once
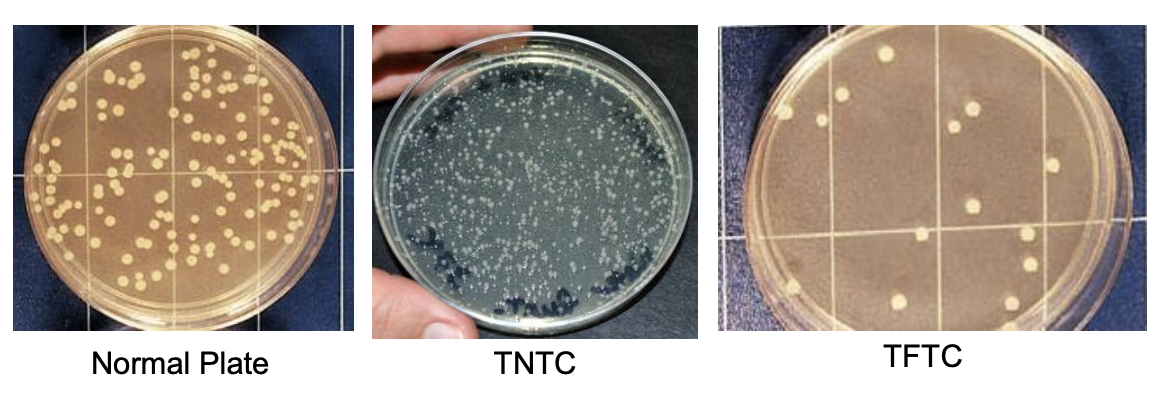
Describe what can is considered a normal plate
Spreader-free plate
Most statistically valid counts
Bacteria = 25 - 250 (30 - 300)
Fungi = 15 - 150
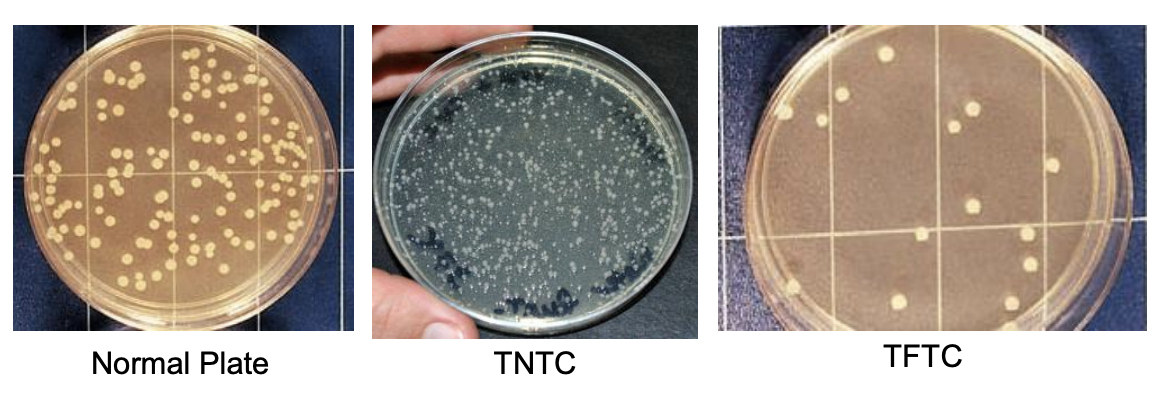
Why do we consider normal plates?
This is because counts outside the normal range may be erroneous in the sense that,
Dilution factors may exaggerate the low counts
Crowded plates may be too difficult to count or may inhibit the growth of some bacteria
6 important rules in calculating and reporting CFU/g or CFU/mL
Compute values from duplicate plates
CFU/mL = liquid samples
CFU/g = solid samples
Report only the first 2 sig figs
Express in scientific notation
Round off to 2 sig figs only at the time of conversion to APC
Explain the rules of rounding off in calculating CFU/mL or CFU/g
Round up when 3rd digit > 5
13,800 = 14 000 = 1.4 ×10^4
Round down when 3rd digit < 5
11,300 = 11,000 = 1.1×10^4
When 3rd digit = 5,
UP if 2nd digit is odd
17,500 = 18,000 = 1.8×10^4
DOWN if 2nd digit is even
18,500 = 18,000 = 1.8×10^4
Explain the formula for computing original cell density if only 1 dilution gives a valid count
Cell density = CFU/mL or CFU/g
CFU/mL or CFU/g = average no. of colonies / original sample volume
Original sample volume = (D) (vol plated)
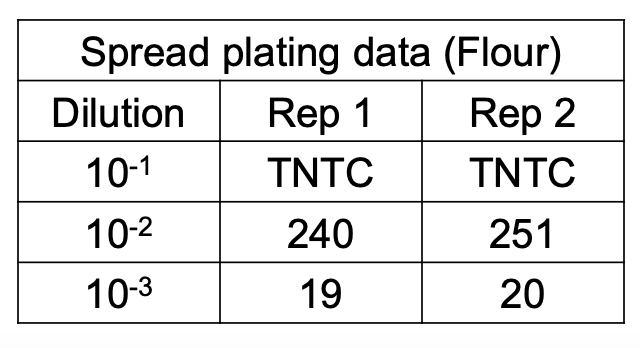
Additional rule for computing original cell density
How do you compute for original cell density if in a duplicate plate set, one falls within the normal range and the other outside?
Solve figure shown
Use only the normal counts
CFU/g = avg no. of colonies / original sample vol
CFU/g = avg no. of colonies / (D) (vol plated)
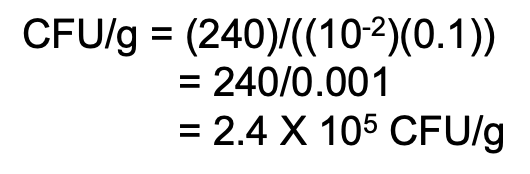
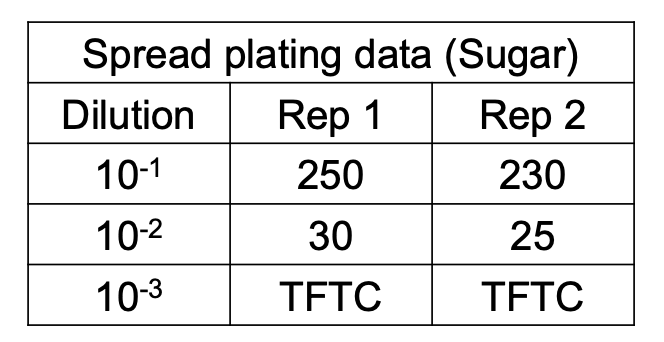
Additional rule for computing original cell density
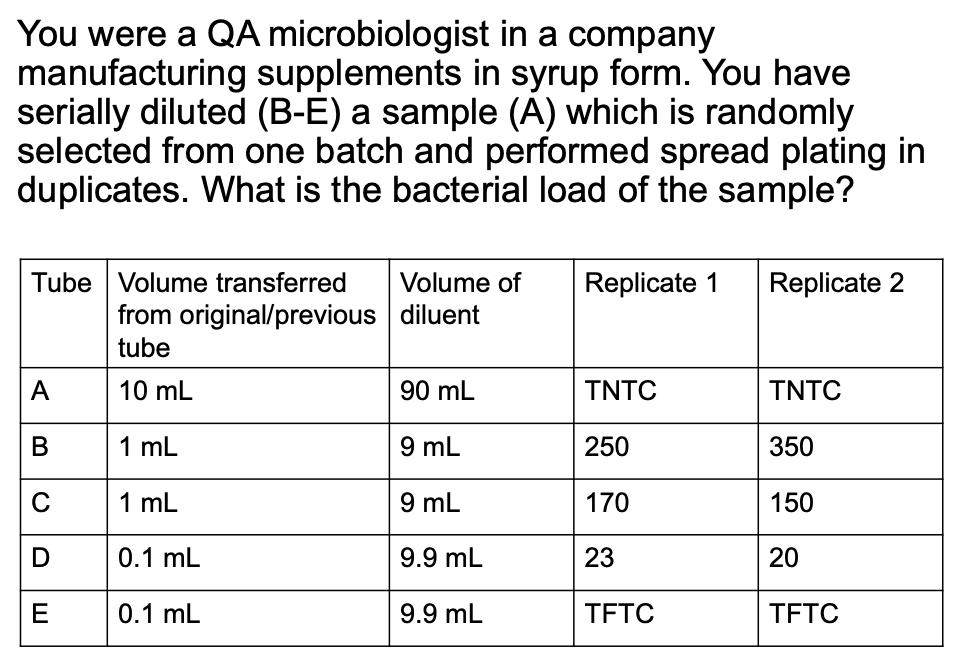
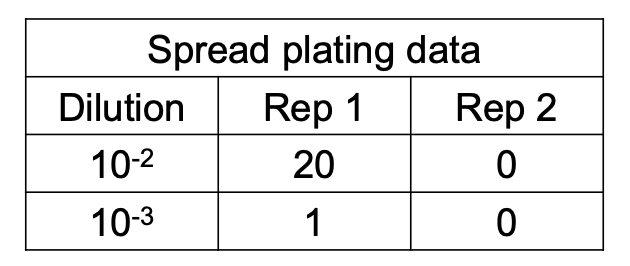
How do you compute for original cell density if 2 consecutive dilutions give a valid count?
Solve figure shown
CFU = total C in plates with valid counts / [(n1 × 1) + (n2 × 0.1)] (D1) (vol plated)
![<ul><li><p>CFU = total C in plates with valid counts / [(n1 × 1) + (n2 × 0.1)] (D1) (vol plated)</p></li></ul><p></p>](https://knowt-user-attachments.s3.amazonaws.com/f22ec3b4-7333-40ef-aea9-bb6ad99089b7.png)
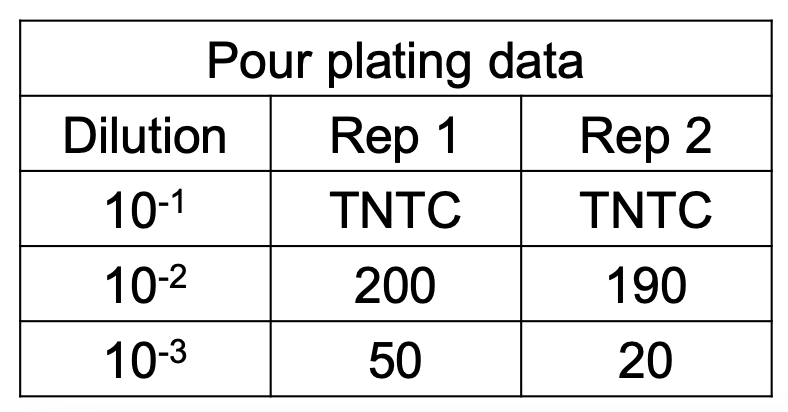
Original cell density
Solve figure shown

Comprehension check
Solve the ff.
_ is reported if there are no normal plates
Estimated Aerobic Plate Count (EAPC)
5 instances when EAPC is used
2n spl
Number of CFU per plate for all dilutions exceeds 250
Number of CFU per plate for all dilutions is less than 25
Spreaders
Plates with no CFU
Laboratory accident
Explain EAPC guidelines when number of CFU per plate for all dilutions exceed 250
Record the counts as TNTC
Count CFU from plate closest to 250
Count CFU in portions of the plate representative of colony distribution
If there are fewer than 100 colonies / cm2
CFU / mL = total no. of colonies in plate / original sample vol
CFU / mL = total no. of colonies in plate / (D) (vol plated)
If there are more than 100 colonies / cm2
CFU / mL = (>100) (DF) (area of the plate)
EAPC Calculation
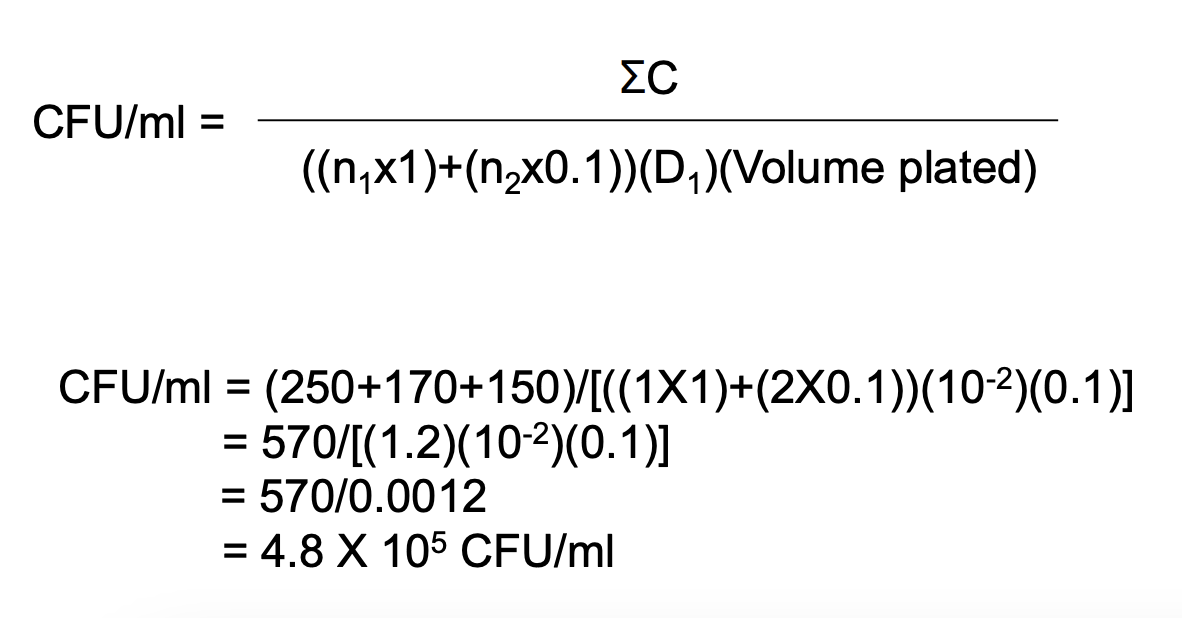
How do you compute for EAPC CFU if the no. of CFU per plate for all dilutions exceed 250 but less than 100 / cm2?
Solve figure shown
EAPC CFU / mL = total no. of colonies in plate / (D) (vol plated)

EAPC Calculation
How do you compute for EAPC CFU if the no. of CFU per plate for all dilutions exceed 250 but more than 100 / cm2?
Solve figure shown
EAPC CFU / mL = >100 (DF) (area of the plate)
area of plate = 65 cm2
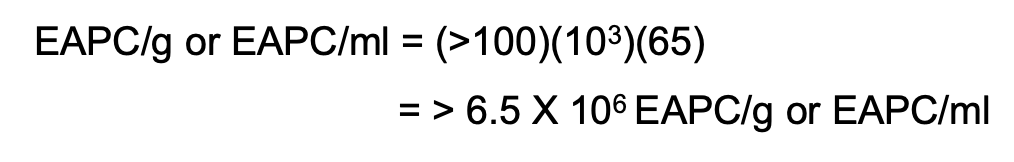
When do we use EAPC computation in the case of spreaders?
When the area covered by spreaders (including total area of repressed growth) exceeds 50% of plate area
When the area of repressed growth exceeds 25% of plate area
Enumerate the different types of spreaders
Chain of colonies formed from disintegration of bacterial clump
Colonies formed in film of water between agar and bottom of plate
Colonies formed in film of water on the edge or at the surface of agar

It must be marked _ to denote estimation from counts outside 25 to 250 per plate range
EAPC
EAPC computation
How do you compute for the EAPC of plates with spreader?
Count each of 3 distinct spreader types as 1 source
If only 1 chain exists, count it as 1 colony
If more than 1 chain appears to originate from separate sources, count each source as 1 colony
Combine spreader count + colony count to compute for EAPC
EAPC computation
How do you compute when all plates yield < 25 CFU or no. of CFU per plate for all dilutions is less than 25?
Solve figure shown
EAPC CFU/mL = < 25 ( 1 / original sample volume)
EAPC CFU/mL = < 25 ( 1 / (D) (vol plated) )
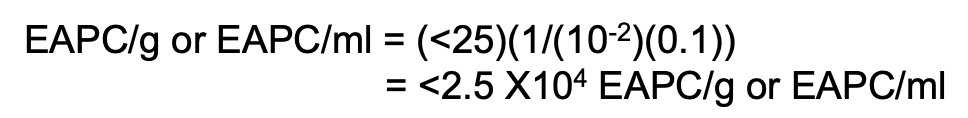
EAPC computation
How do you compute for EAPC when all plates yield no CFU or all plates from all dilutions have no colonies?
EAPC CFU/mL = < 1 (lowest dilution used)
When plates are known to be contaminated or unsatisfactory, mark it as _ to denote laboratory accident
EAPC
Properties of a good diluent for homogenizing food samples
onn
Osmotic balance = should match the osmotic pressure of microbial cells to prevent cell lysis or dehydration
Non-toxic to microorganisms = should not inhibit microbial growth to allow isolation, enumeration
No or minimal nutritional value = should not support microbial growth before plating, to prevent overgrowth