BIOL 3301: Ch 15 Gene Regulation in EUkaryotes I: General Features of Transcriptional Regulation
1/34
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
35 Terms
gene expression
process by which the information within a gene is used to synthesize RNA and polypeptides, and to affect the properties of cells and the phenotype of multicellular organisms
gene regulation
the level of gene expression can vary under different conditions
importance of gene regulation
respond to environmental changes such as nutrient availability, stress, etc.
produce different cell types in multicellular species
facilitate changes during development
some genes are only expressed during embryonic stages, whereas others are only expressed in the adult
eukaryotic regulation of gene expression
transcription
regulatory transcription factors- activate or inhibit transcription
activators or repressors
combinatorial control
most eukaryotic genes are regulated by many factors
common factors contributing to ? are
1 or more activator proteins may stimulate transcription
1 or more repressor proteins may inhibit transcription
activators and repressors may be modulated by binding of small effector molecules, protein-protein interactions, and covalent modifications
transcription factors
proteins that influence the ability of RNA polymerase to transcribe a given gene
2 main types:
general transcription factors
required for RNA pol to bind to the core promoter and for its progression to the elongation stage
necessary for basal transcription < very low transcription rate, usually can’t even be detected
regulatory transcription factors
serve to regulate transcription rate of target genes
they influence the ability of RNA pol to begin the transcription of a particular gene
general transcription factors
required for RNA pol to bind to the core promoter and for its progression to the elongation stage
necessary for basal transcription < very low transcription rate, usually can’t even be detected
regulatory transcription factors
serve to regulate transcription rate of target genes
they influence the ability of RNA pol to begin the transcription of a particular gene
domains
regions in transcription factor proteins that have specific functions
DNA binding
binding site for small effector molecules
dimerization
ex., in many proteins, two identical subunits can bind to each other to form a functional dimer.
motif
a domain, or a portion of a domain that has a very similar structure in many different proteins
a polypeptide sequence that binds DNA that has a particular fx (e.g., DNA binding ? to help the protein grab on to the DNA).
domain
regions in transcription factor proteins that have specific functions: DNA binding, binding site for small effector molecules, dimerization
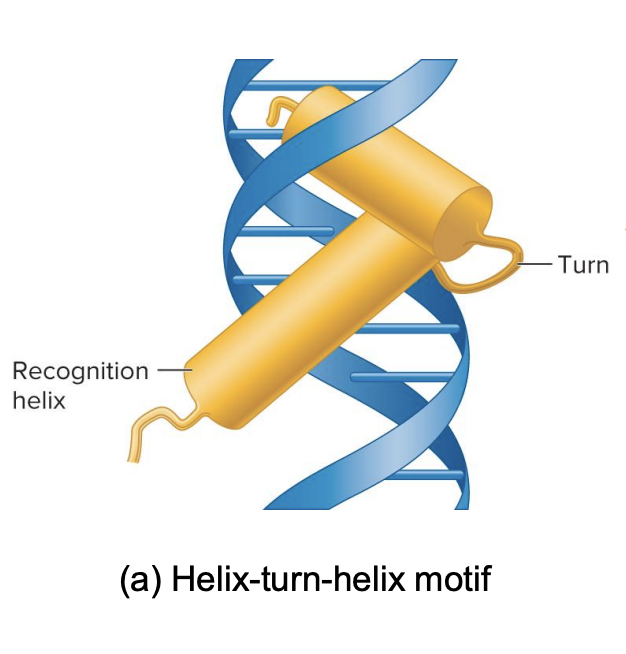
helix turn helix motif
The motifs are special shapes or patterns in the transcription factors that allow them to bind to the DNA
It’s like two sticks (α-helices) connected by a short bend (turn). The sticks help the protein bind to DNA.
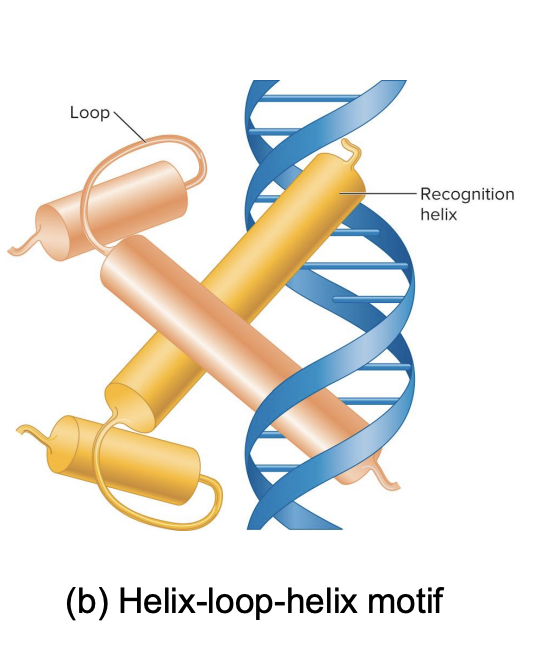
helix loop helix motif
The motifs are special shapes or patterns in the transcription factors that allow them to attach to the DNA.
This motif looks like a small loop connecting two sticks. Two of these motifs can pair up and help the protein grab onto DNA.
regulatory domain
in transcription factors, the ? can bind to specific DNA sequences or to other proteins (co-factors) to either activate or repress the transcription of genes
regulatory elements, control elements, regulatory sequences
DNA sequences within enhancers that control gene activity by binding to regulatory transcription factors (activators or repressors)
(activators/repressors) regulatory transcription factors binding to ? affects the (increase/decrease) transcription of an associated gene
cis regulatory elements
enhancer
a DNA region, usually 50-1000 bp in length, that contains one or more of the regulatory elements/control elements/regulatory sequences
activator
regulatory protein (regulatory transcription factor) that increases the rate of transcription
binds to regulatory element in enhancer
repressor
regulatory protein/regulatory transcription factor that decreases the rate of transcription
binds to regulatory element in enhancer
activators
proteins (regulatory transcription factor) bind to regulatory elements (enhancers) far from the gene's core promoter.
DNA loops bring the activator close to the core promoter and preinitiation complex.
This interaction increases RNA transcription by helping the transcription machinery start.
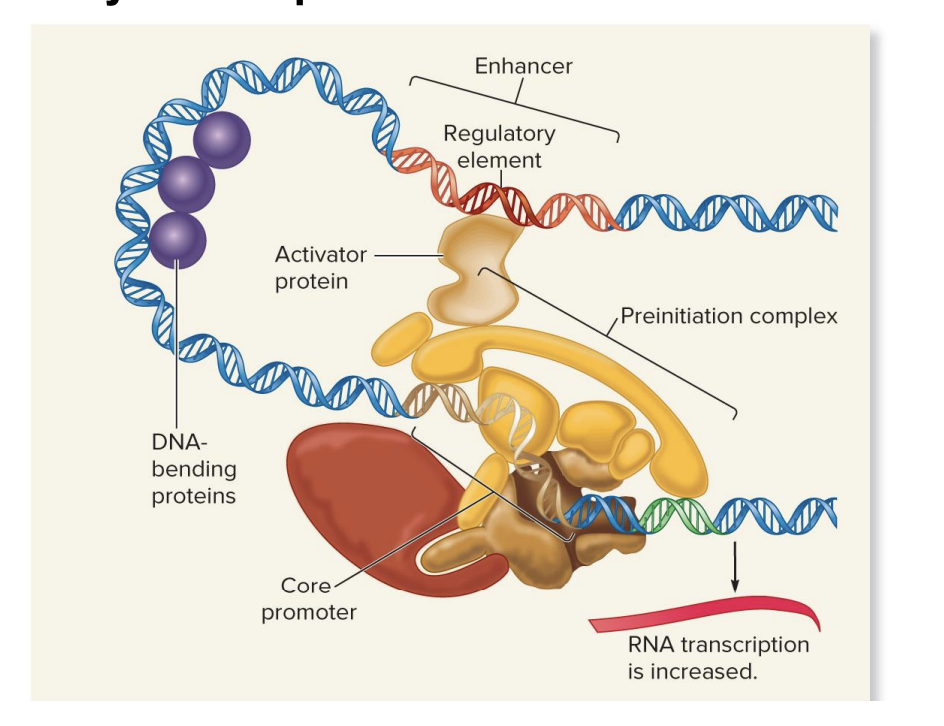
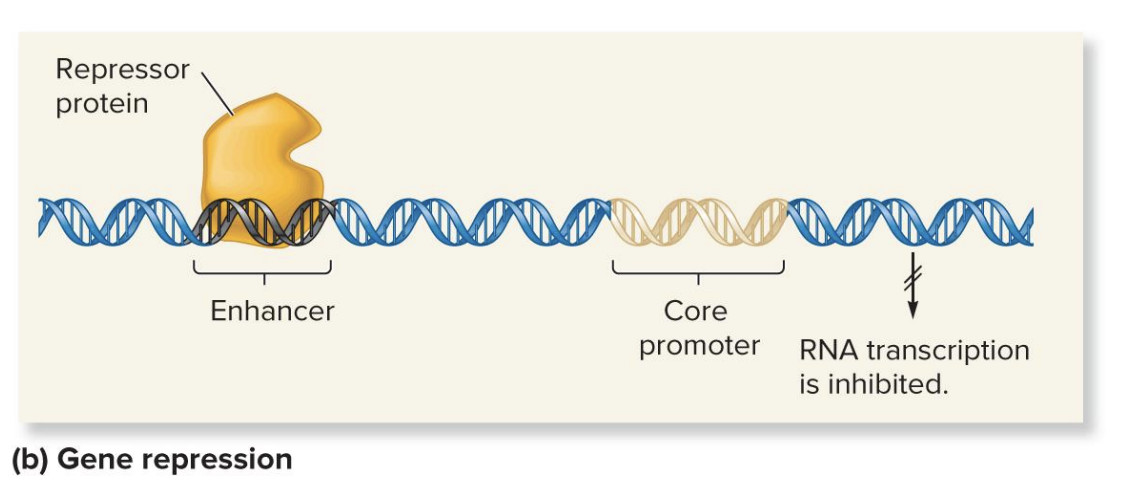
repressors
proteins (regulatory transcription factor) can bind to enhancers and block the preinitiation complex.
This prevents the transcription machinery from starting or completing RNA transcription, thus inhibiting transcription.
up regulation
can be 10 to 1000 fold
the binding of an activator (regulatory transcription factor) to an enhancer increases the rate of transcription
down regulation
binding of a repressor (regulatory transcription factor) that inhibits transcription
enhancers
orientation independent/bidirection
can function in forward (5’→3’) or reverse (3’→5’) orientation
location
some can be close to the promoter, just a few hundred nucleotides upstream
However, they are often further away from promoter, 1000s or even 100,000 nucleotides away from the gene they regulate.
Enhancers can be downstream from the promoter or even within introns of the gene.
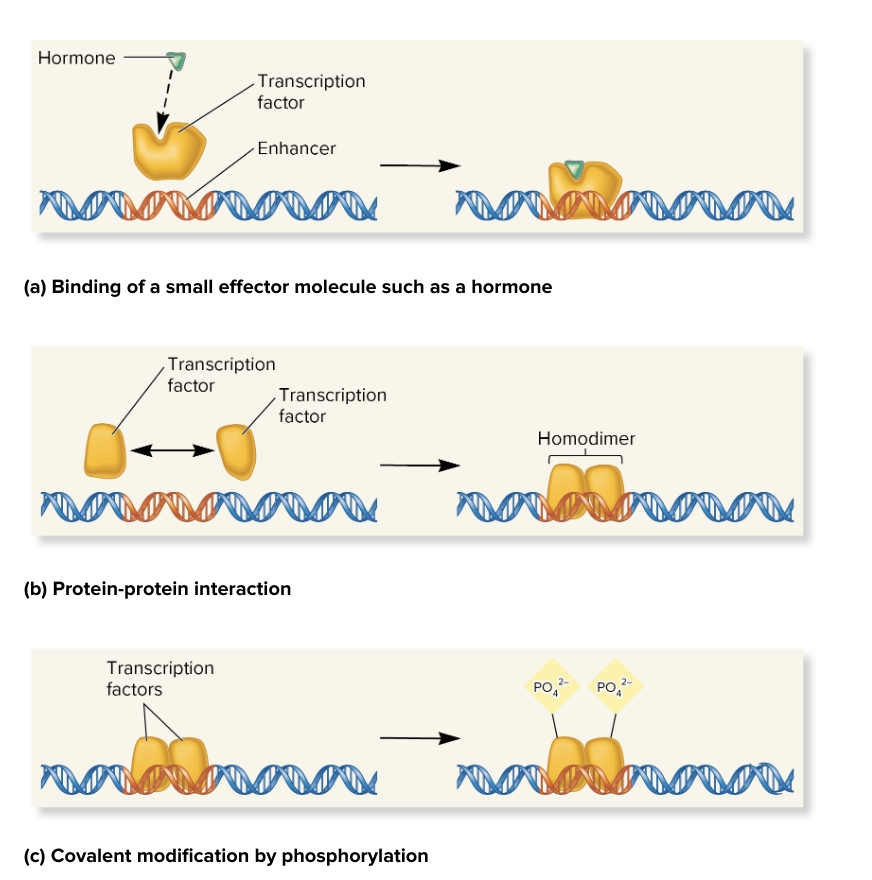
ways to modulate fx of regulatory transcription factors
binding a small effector molecule (e.g., hormone)
small molecule binds RTF, changing its ability to bind DNA
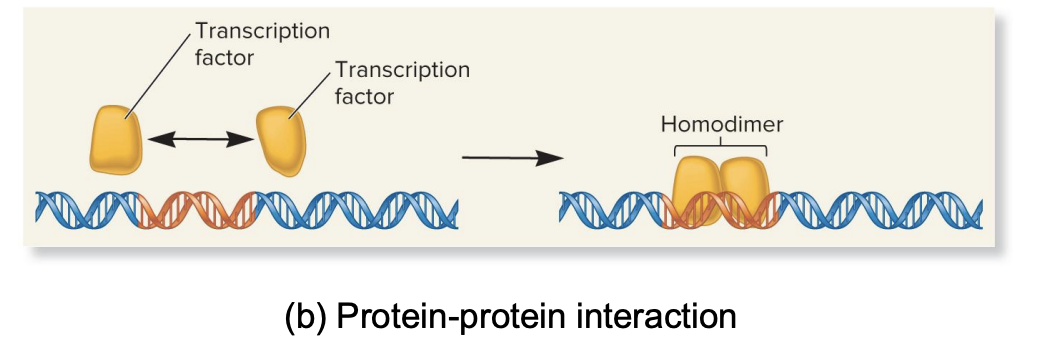
protein-protein interaction
RTF can form interactions with each other (e.g., dimerization), influencing their fx and DNA binding
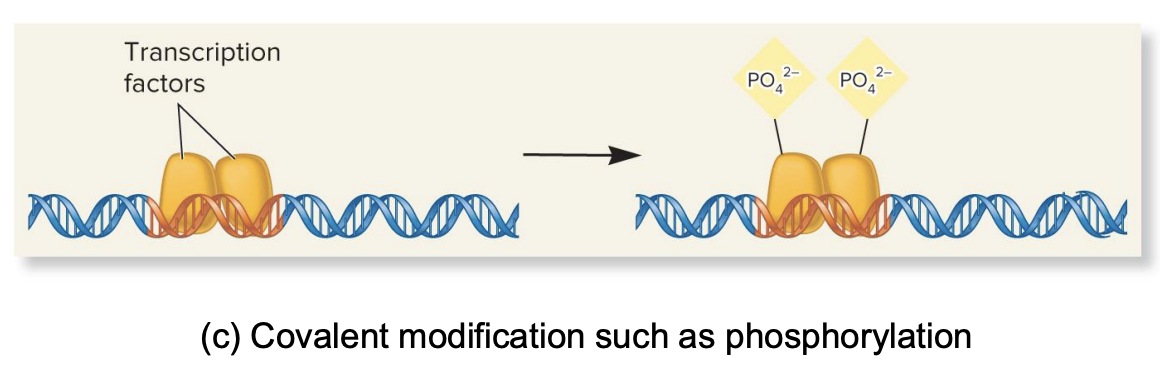
covalent modifications (e.g., phosphorylation)
chemical changes like phosphorylation (adding phosphate groups) can alter the fx of transcription factors
e.g., When it is phosphorylated, it can bind to DNA and activate transcription
RTF binds small effector molecule
binding a ? (e.g., hormone)
small molecule binds RTF, changing its ability to bind DNA
way to modulate fx of regulatory transcription factors
protein-protein interaction
RTF can form interactions with each other (e.g., dimerization), influencing their fx and DNA binding
ways to modulate fx of regulatory transcription factors
covalent modification
chemical changes like phosphorylation (adding phosphate groups) can alter the fx of transcription factors
e.g., When it is phosphorylated, it can bind to DNA and activate transcription
ways to modulate fx of regulatory transcription factors
steroid receptors
regulatory transcription factors that respond to steroid hormones
steroid hormone binds the regulatory transcription factor
ultimate action of steroid hormone is to affect gene transcription
steroid hormones
these hormones bind to the regulatory transcription factor
purpose is to affect gene transcription
secreted by endocrine glands into the bloodstream
then taken up by cells that respond to the hormone
cells respond to ? hormones in different ways
glucocorticoids (? hormone)
influence nutrient metabolism in most cells
promote glucose utilization, fat mobilization, and protein breakdown
glucocorticoid response elements GRE
DNA sequences within enhancers
located near dozens of different genes, allowing the hormone (glucocorticoid) to activate many genes
consists of 2 sequences that are close together
5’ AGRACA 3’
3’ TCYTGT 5’
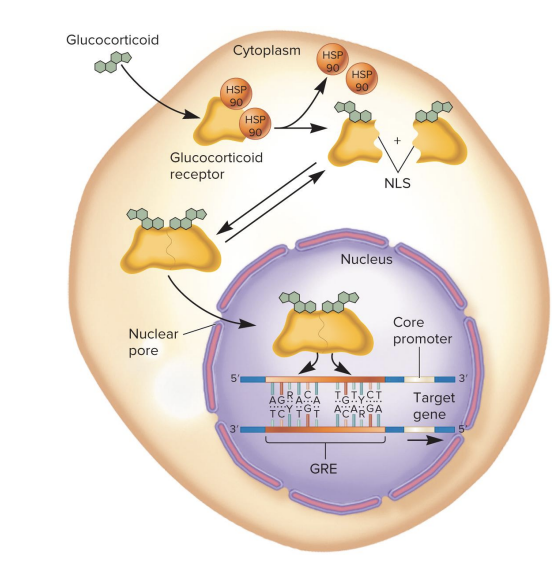
glucocorticoid hormones enter cell and bind to the glucocorticoid receptor subunits that dimerize, enter the nucleus, bind to ? and activate gene transcription
glucocorticoids
steroid hormones
influence nutrient metabolism in most cells
promote glucose utilization, fat mobilization, and protein breakdown
enter cell and bind to the glucocorticoid receptor subunits that dimerize, enter the nucleus, bind to a Glucocorticoid Response Element (GRE) and activate gene transcription
glucocorticoid hormone action
Glucocorticoid hormone enters the cell and binds to the glucocorticoid receptor.
The binding of the hormone causes the release of HSP90 (a heat shock protein).
This release exposes a nuclear localization signal (NLS) on the receptor.
The two glucocorticoid receptors with the hormone dimerize and enter the nucleus.
Inside the nucleus, the dimer binds to the Glucocorticoid Response Element (GRE) on the DNA.
The binding of the receptors to the GRE activates transcription of a nearby gene.
GRE can be located far away from the target gene, even within an enhancer.
CREB cAMP response element-binding protein
a regulatory transcription factor
gets activated when cAMP (cyclic adenosine monophosphate) levels increase inside the cell
cAMP is a common signaling molecule
extracellular signaling molecules cause an increase in cytoplasmic [cAMP]
binds to DNA at two adjacent sites called CREs (cAMP Response elements)
CRE consensus sequence
5’-TGACGTCA-3’
3’-ACTGCAGT-5’
binding to the CRE allows ? to help activate transcription of target genes
cAMP response element CRE
consensus DNA sequence
5’-TGACGTCA-3’
3’-ACTGCAGT-5’
binds CREB protein, a transcription factor
CREB activated when cAMP levels rise
activated CREB boosts gene transcription
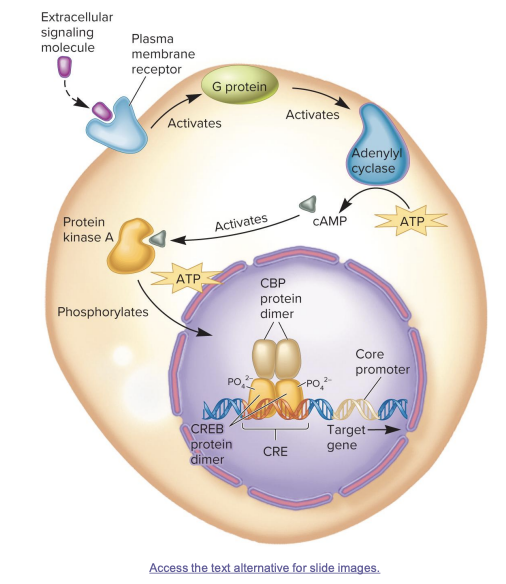
CREB protein action
extracellular signaling molecules (like epinephrine) trigger cAMP production inside the cell
cAMP acts as a second messenger
First messenger = signal from outside the cell (like a hormone)
Second messenger = small molecule inside the cell (like cAMP) that carries the message and activates cellular responses.
cAMP activates protein kinase A (PKA)
PKA phosphorylates CREB protein, activating it
phosphorylated CREB binds co-activator CREB-binding protein CBP→ greatly increase adjacent gene transcription
CREB protein pathway to transcriptional activation
A signal molecule (like a hormone) binds to a membrane receptor, activating a G protein.
The G protein activates adenylyl cyclase, which makes cAMP from ATP.
cAMP activates protein kinase A (PKA).
PKA enters the nucleus and phosphorylates the CREB protein.
Phosphorylated CREB binds to CBP (a coactivator) and activates transcription of the target gene at the CRE site.