Nucleic Acids and DNA Replication
1/61
Earn XP
Description and Tags
macromolecules III
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
62 Terms
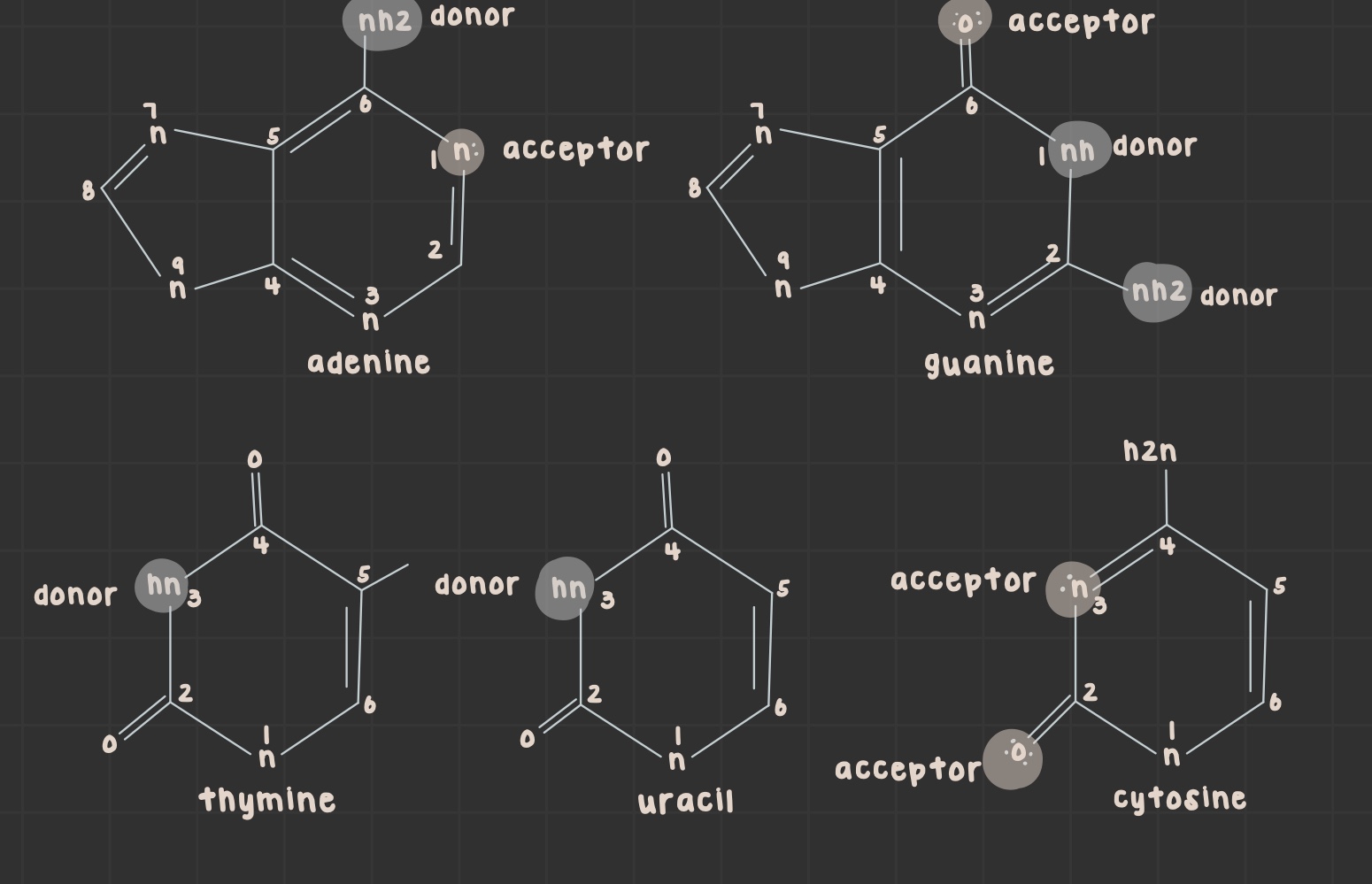
What is a purine?
Nitrogenous base with two rings (adenine and guanine)
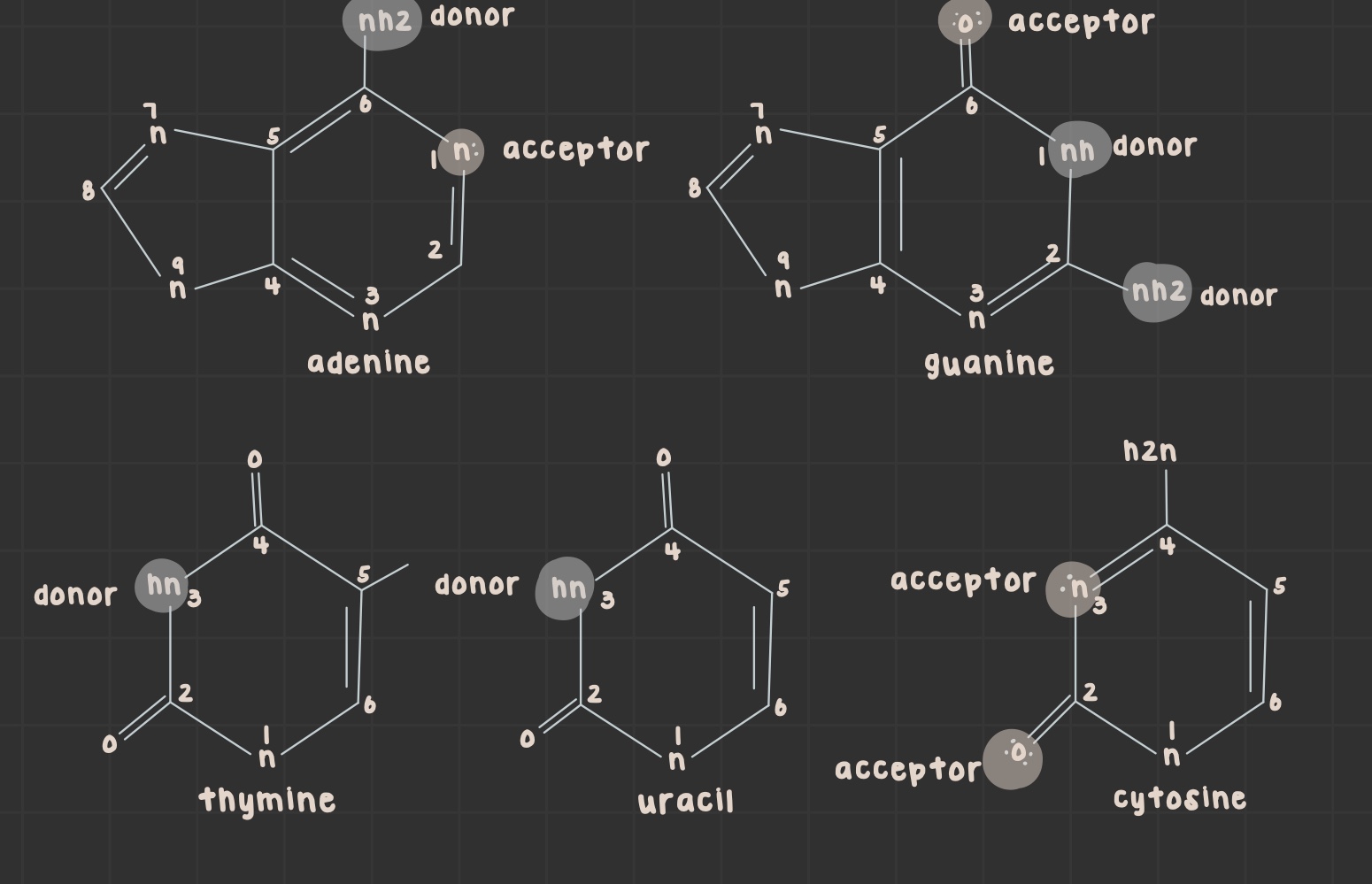
What is a pyrimidine?
Nitrogenous base with one ring (uracil, thymine, and cytosine)
What is tautomerization?
proton transfer from one site within a molecule to another site in the same molecule.
What is deamination?
Amino groups within adenine, guanine and cytosine can be removed and replaced spontaneously by carbonyls
What happens during cytosine deamination?
cytosine replaces its amino group with a carbonyl and is converted to uracil (error in DNA)
What is methylation?
Cytosine can become methylated at position 5 to alter DNA expression
What happens to cytosine after it is methylated and deaminated?
it is converted to thymine, potentially leading to mutations.
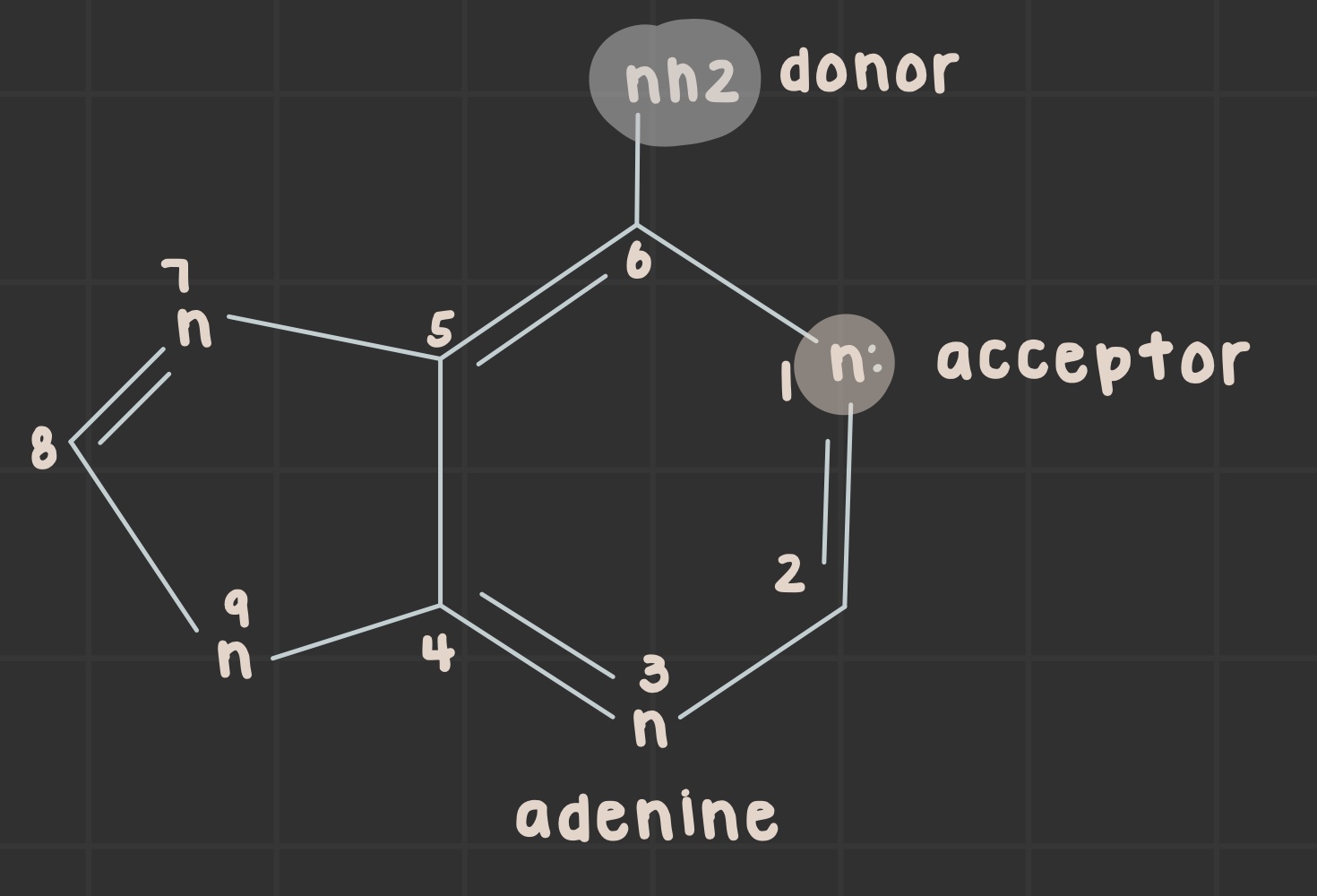
What is the structure of adenine?
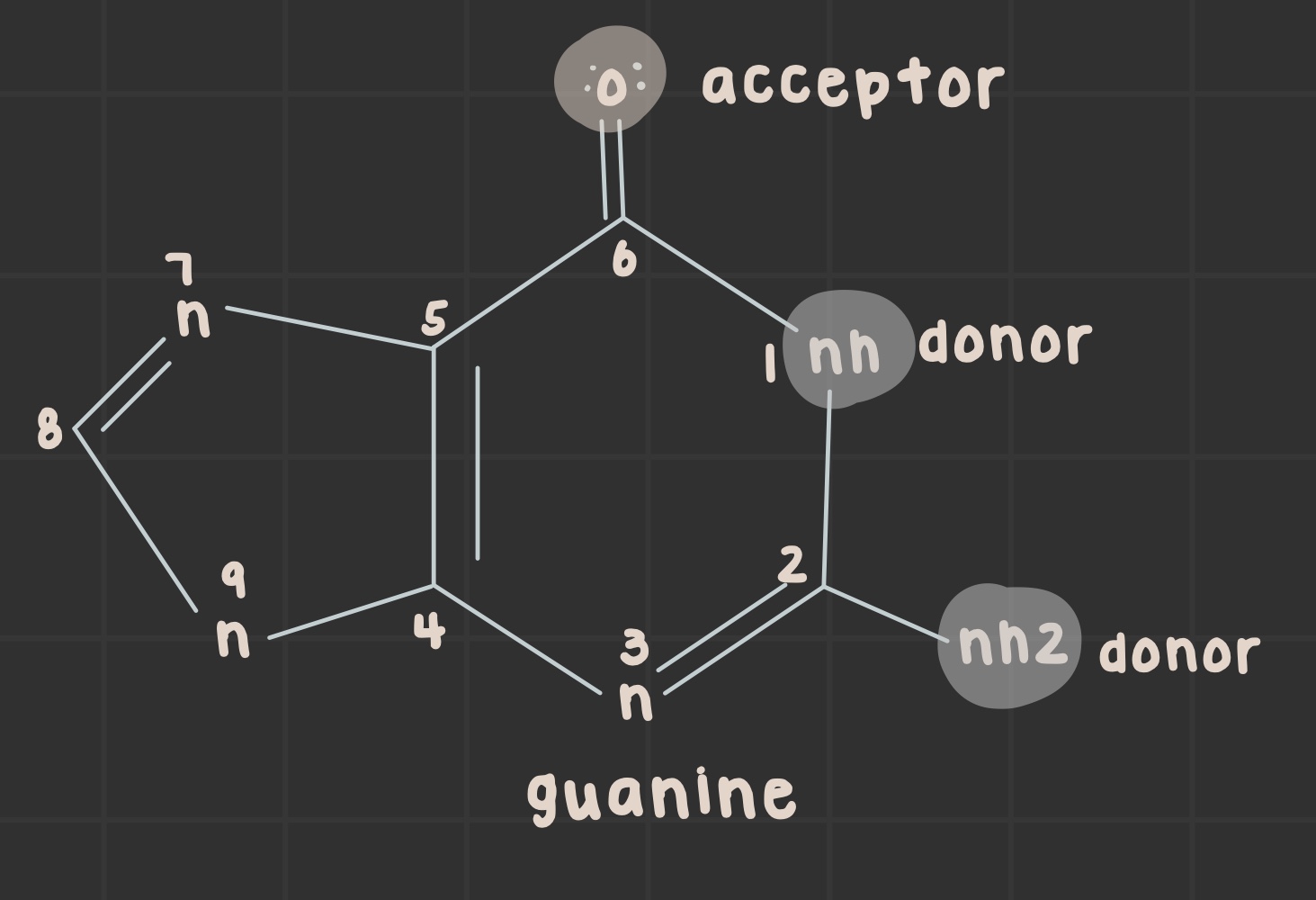
What is the structure of guanine?
What is the structure of thymine?
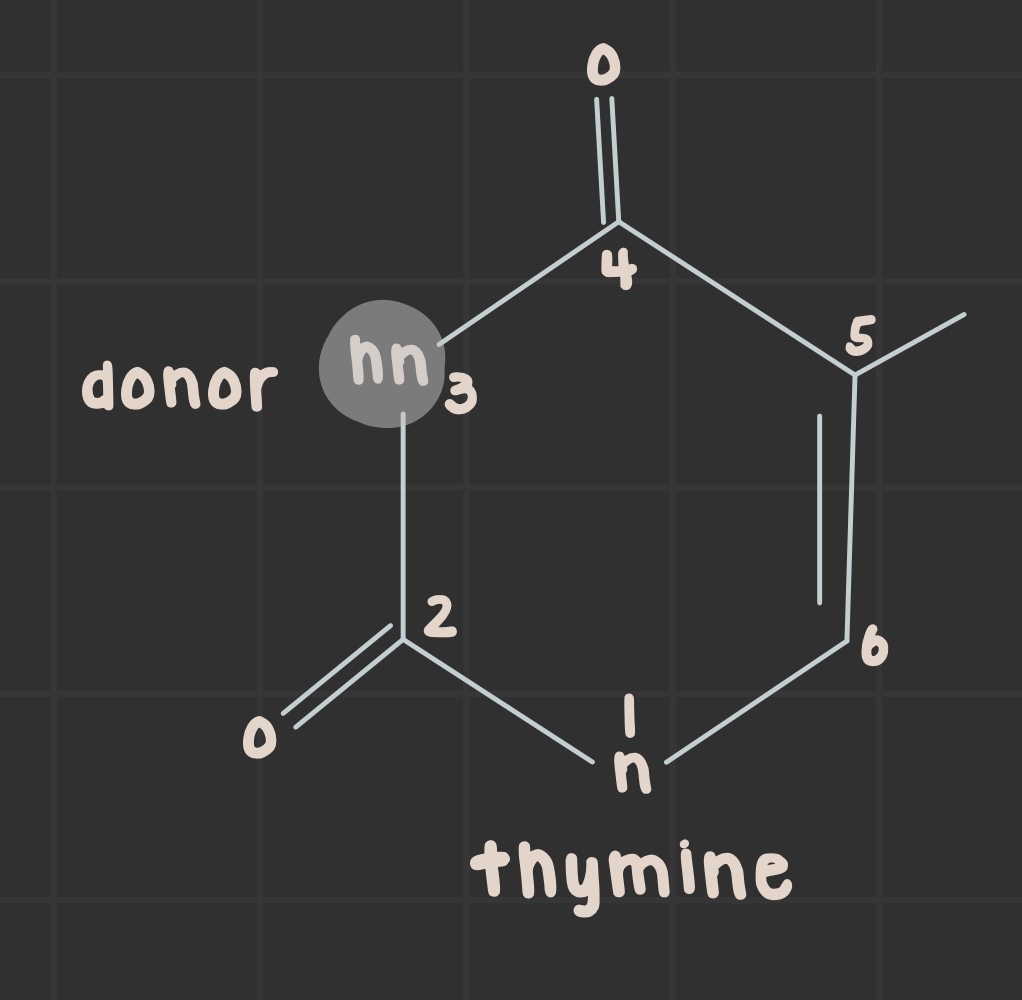
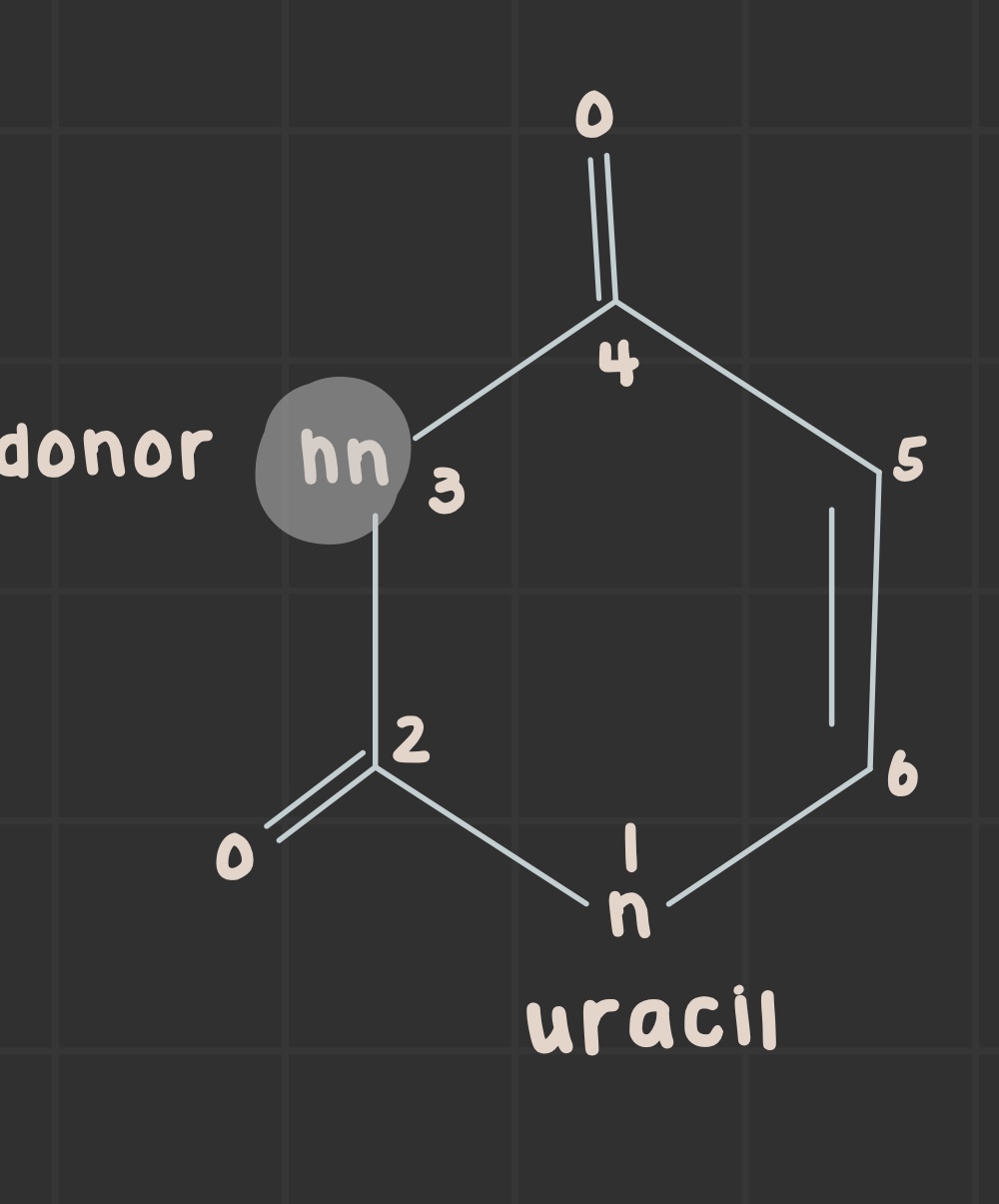
What is the structure of uracil?
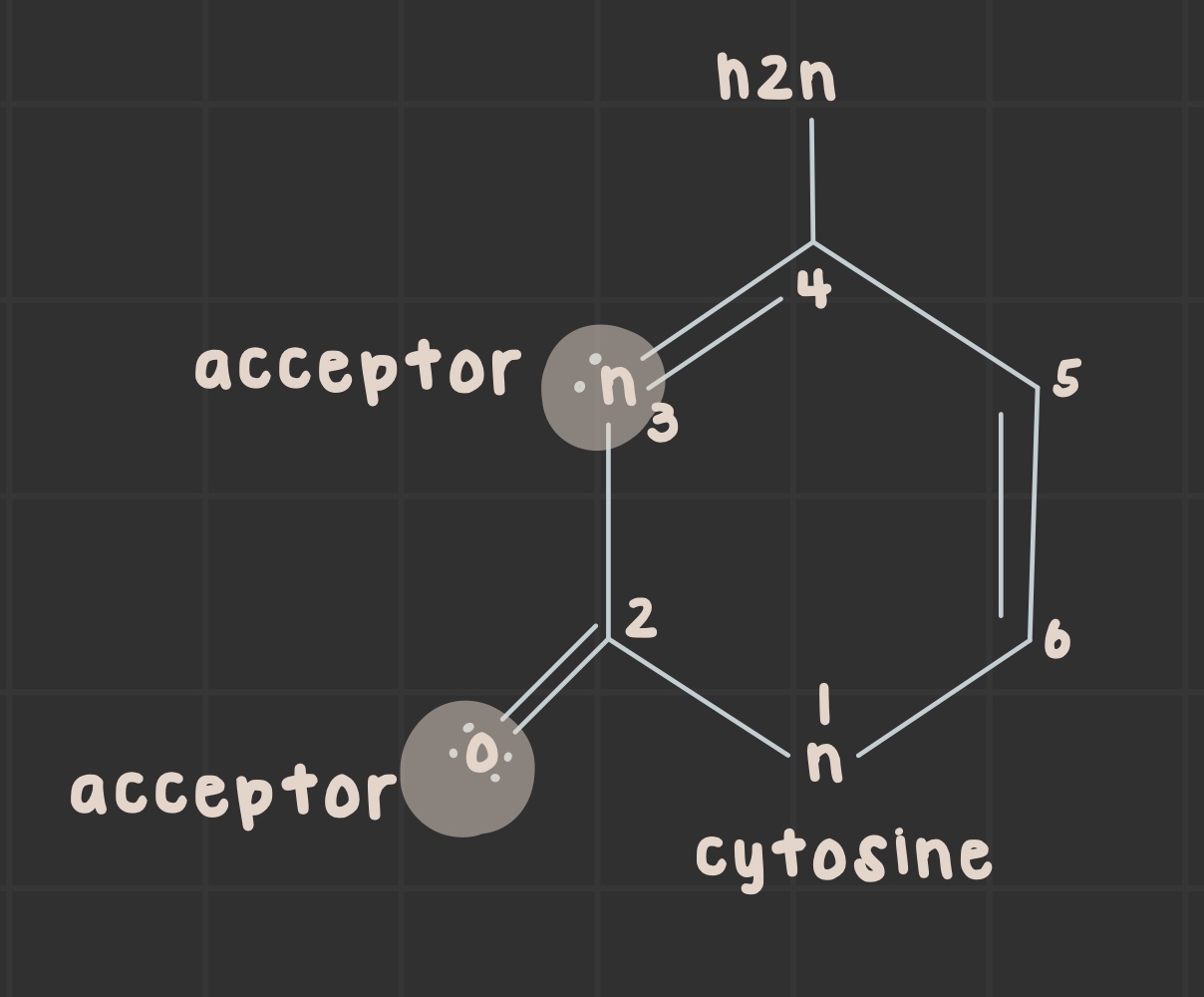
What is the structure of cytosine?
What is DNA?
Two antiparallel nucleic acid strands of deoxyribonucleotides and deoxyribose sugar wrapped in a double helix that transmits genetic information
What is a nucleotide?
Nitrogenous base, sugar, and 1-3 negatively charged phosphates.
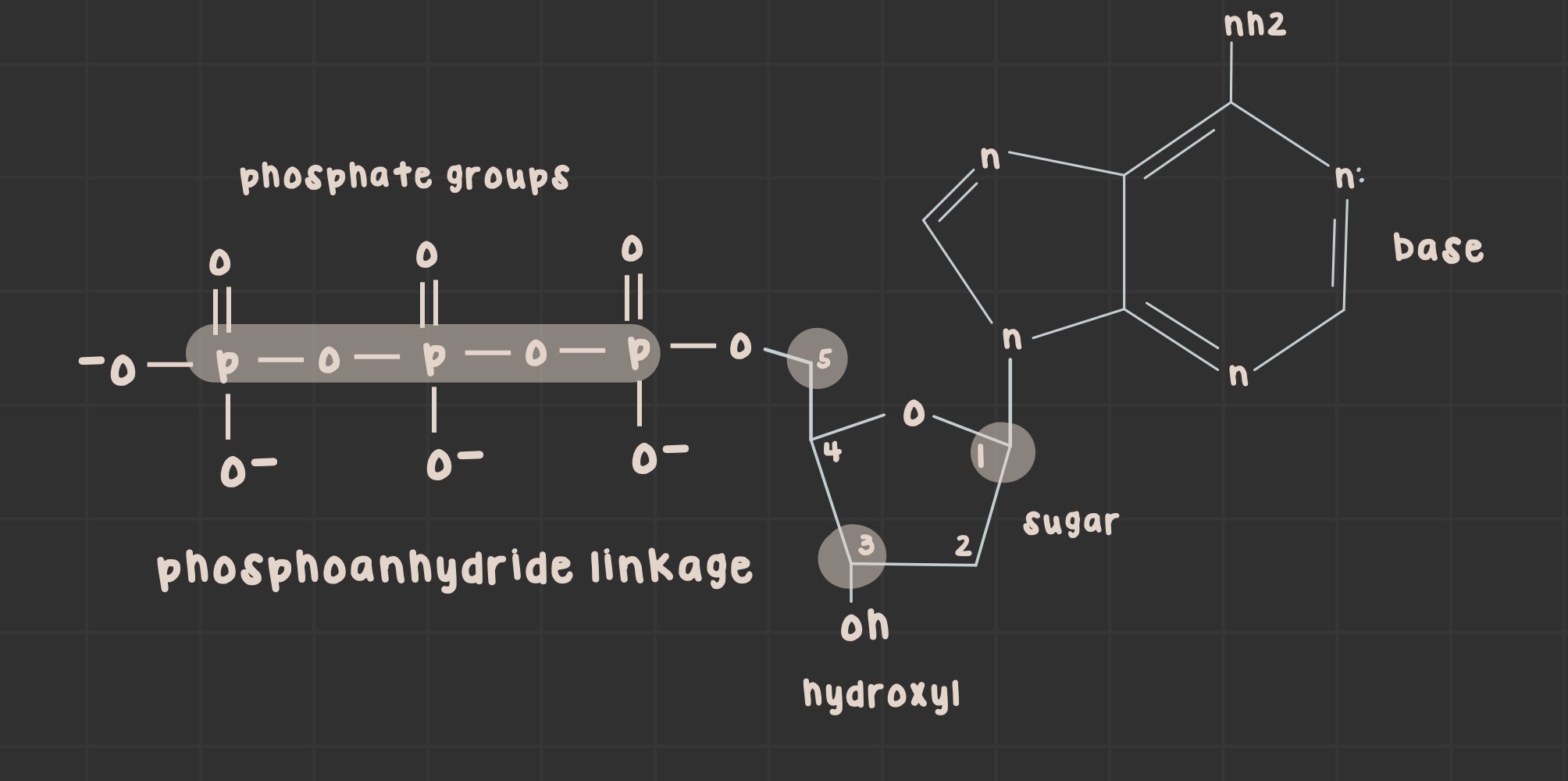
What is a nucleoside?
Nitrogenous base and sugar linked by a glycosidic bond.
pyrimidines: connect C1 to N1
purines: connect C1 to N9
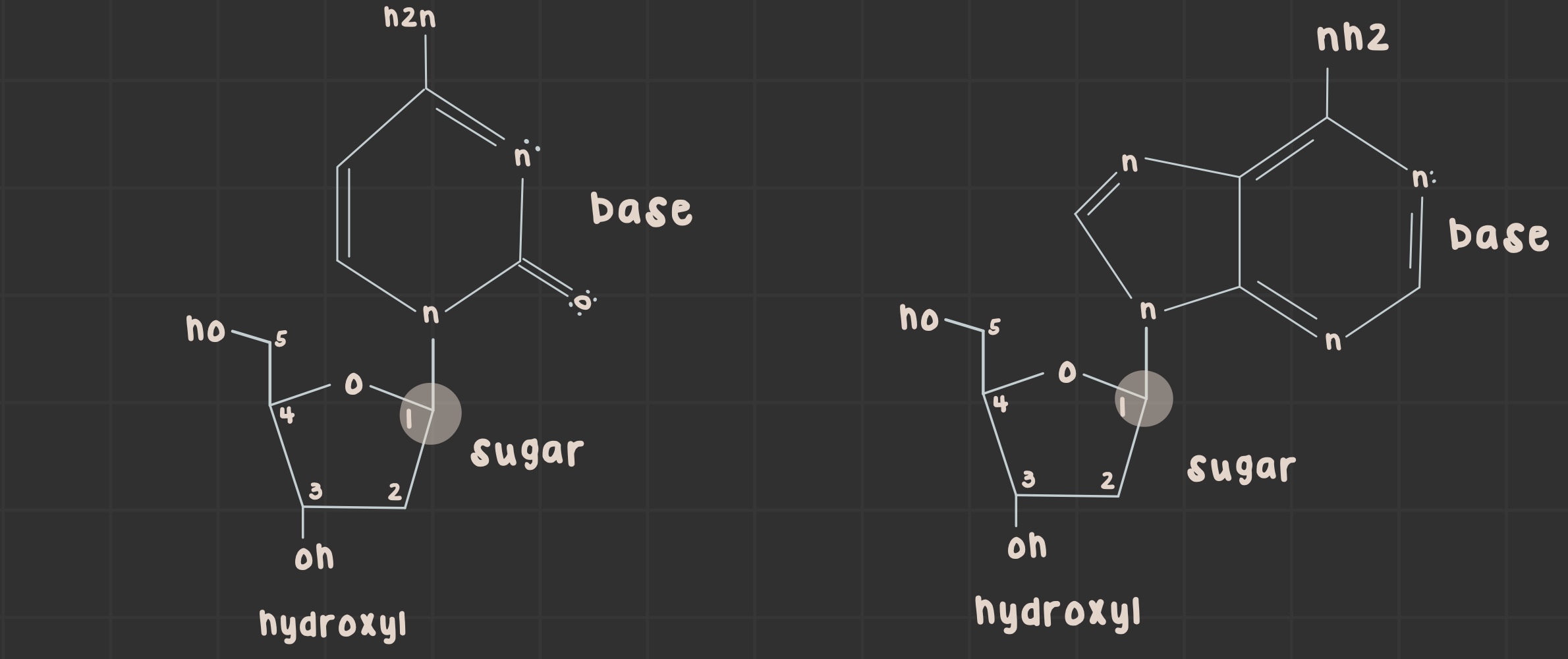
What is a nucleic acid?
Nucleotide polymer connected by covalent phosphodiester bonds (written from 5' to 3')
What is hybridization?
Joins two nucleic acid strands via a double helix.
What is denaturation?
Disruption of hydrogen bonds/base interactions (loss of 2° structure) via high temperature, abnormal pH, and changes to salt concentration
What is reannealing?
restranding of DNA that occurs when solution temperature is decreased below Tm.
What is DNA replication?
Synthesis of DNA during the S phase of the cell cycle.
What is semiconservative replication?
One new daughter strand is synthesized via the parent template
What is the origin of replication?
Beginning of replication; marked by specific nucleotide sequences in DNA.
enzymes attach, strands separate, and a replication bubble forms where DNA replication occurs in both directions
How is DNA antiparallel?
The 3' end of one strand aligns with the 5' end of the other
What is Chargaff's rule in DNA?
Equal numbers of A-T and G-C hydrogen bonds between strands.
What is B-DNA?
Right-handed helix, turns every 3.4 nm (~10 bases), with major and minor grooves (most common)
What is Z-DNA?
Left-handed helix, turns every 4.6 nm (~12 bases), found in GC-rich DNA or high salinity DNA (uncommon/unstable)
What is the leading strand?
Continuously synthesizes DNA toward the replication fork; 3’-5’
What is the lagging strand?
Synthesized discontinuously in a direction away from the replication fork; 5’-3’
What are Okazaki fragments?
Short DNA fragments formed from RNA primers being periodically added on the lagging strand while DNA Pol synthesizes.
What are telomeres?
Noncoding DNA sequences (5'-TTAGGG-3') added to the end of linear chromosomes that prevents a loss of genetic information
What is telomerase?
Replenishes telomere sequences; overexpression is linked to cancer.
What is the Hayflick limit?
Number of times a cell replicates before telomeres are too short for division.
What is senescence?
Growth arrest due to telomeres shortening past a critical point (i.e., aging).
What is base pair mismatch?
DNA pol adds incorrect nucleotides into the new strand during replication (S phase) causing mismatch between the parent and daughter strands.
DNA pol uses exonuclease activity to replace incorrectly paired nucleotides
What is mismatch repair (MMR)?
An incorrect base pair is detected after replication (G2) and causes single-stranded breaks in the new strand.
endonucleases remove the mismatched base and several nucleotides on either side from the strand
phosphodiesterase bonds are cleaved in the middle of the strand
DNA pol adds the correct nucleotides after incorrect ones are removed
DNA ligase forms new phosphodiester bonds to rejoin the strands
What is base excision repair?
Corrects damage that occurs during the cell cycle (G1/G2) that does not significantly distort the helix (oxidation, deamination, and alkylation of bases)
glycosylase: recognizes the site of DNA damage and excises the damaged base
endonuclease: cleaves the phosphodiester bond
DNA pol: fills the gaps
DNA ligase: rejoins DNA strands
What are mutagens?
Chemical, physical, or biological agents that can lead to mutation.
What are carcinogens?
Agents that lead to cancer-causing mutations.
What are chromosomal mutations and examples?
Mutations over a large segment of DNA; disrupt function of numerous genes.
deletion: DNA removed
duplication: DNA repeated
inversion: DNA breaks off and reattaches in reverse orientation
translocation: DNA breaks off and attaches to another chromosome
What is deletion?
DNA removed.
What is duplication?
DNA repeated.
What is inversion?
DNA breaks off and reattaches in the reverse orientation.
What is translocation?
DNA breaks off and attaches to another chromosome.
What is prokaryotic DNA?
Single circular chromosome.
What is eukaryotic DNA?
Multiple linear chromosomes
coding: DNA sequences (genes) with information needed for protein production/cell function
noncoding: maintenance of chromosomal integrity (telomeres) or gene expression regulation
What is homologous recombination?
If there is a diploid organism/sister chromatid nearby, the chromosome acts as a template to repair the broken chromosome (double-stranded breaks)
What is nonhomologous end joining?
If there is a haploid organism/no duplicated chromosome, broken chromosomes are rejoined by enzymes.
What is a nucleosome?
DNA helix wraps around 8 histone proteins twice during early chromatin formation
histone types: h1, h2a, h3, h4
DNA wrapped octamer core: 8 subunits (h2a, h2b, h3, h4 ×2)
h1 (linked histone): present outside core
What is nucleotide excision repair?
Corrects extensive/bulky damage to DNA (UV radiation causing thymine dimers which distort the helix) that occurs during the cell cycle (G1/G2)
NER endonuclease: removes the damaged region
DNA pol: fills the gaps
DNA ligase: rejoins repaired strands
What is heterochromatin?
DNA tightly coiled around histone proteins (ionic interactions); telomeres and centromeres.
What is euchromatin?
Histones are modified via acetylation of K residues to neutralize K's positive charge.
What are phosphoanhydride bonds?
bonds that link ATP phosphate groups together
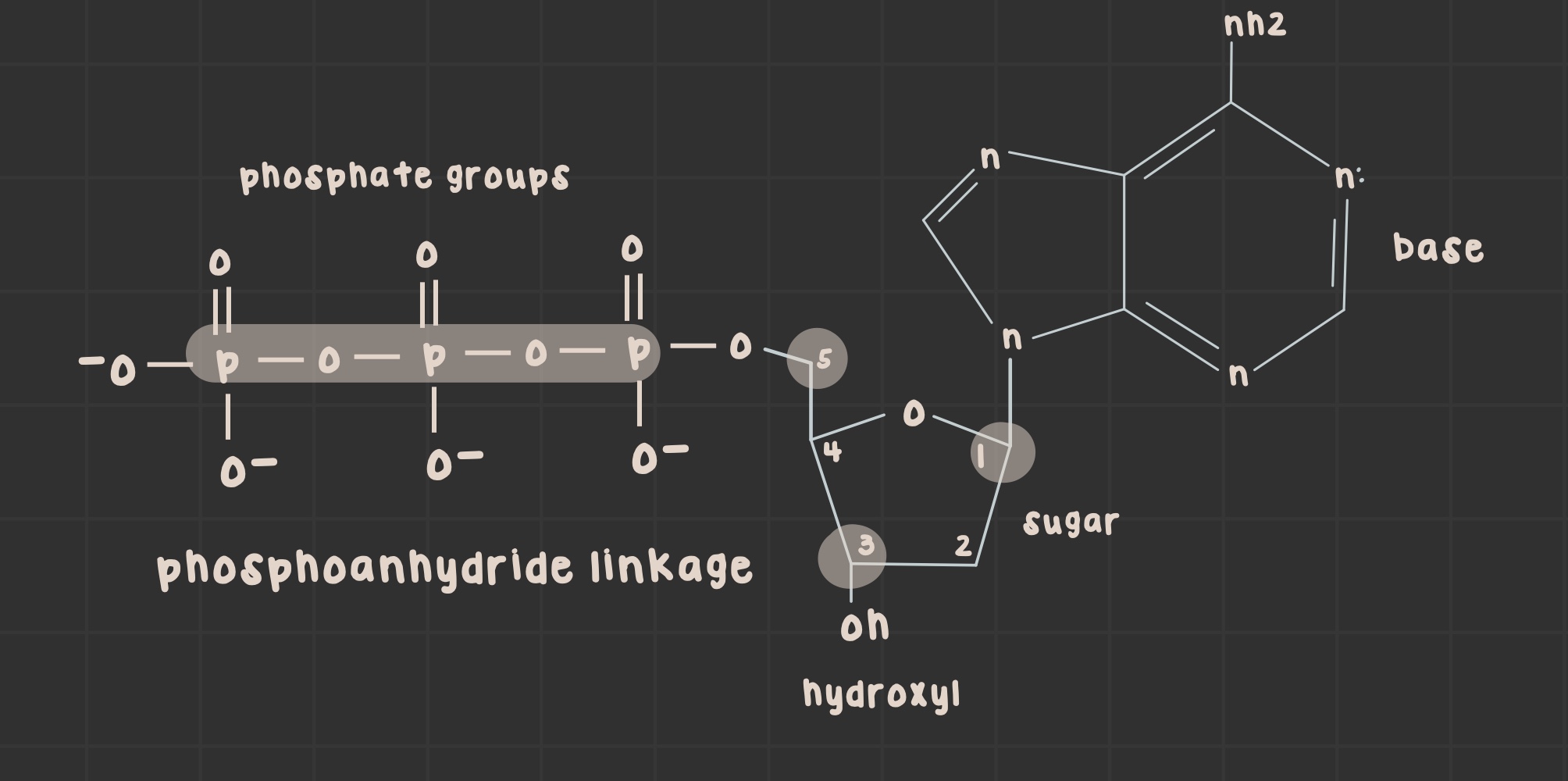
Define dNTP.
deoxynucleotide triphosphates (nitrogenous base + 3 phosphates)
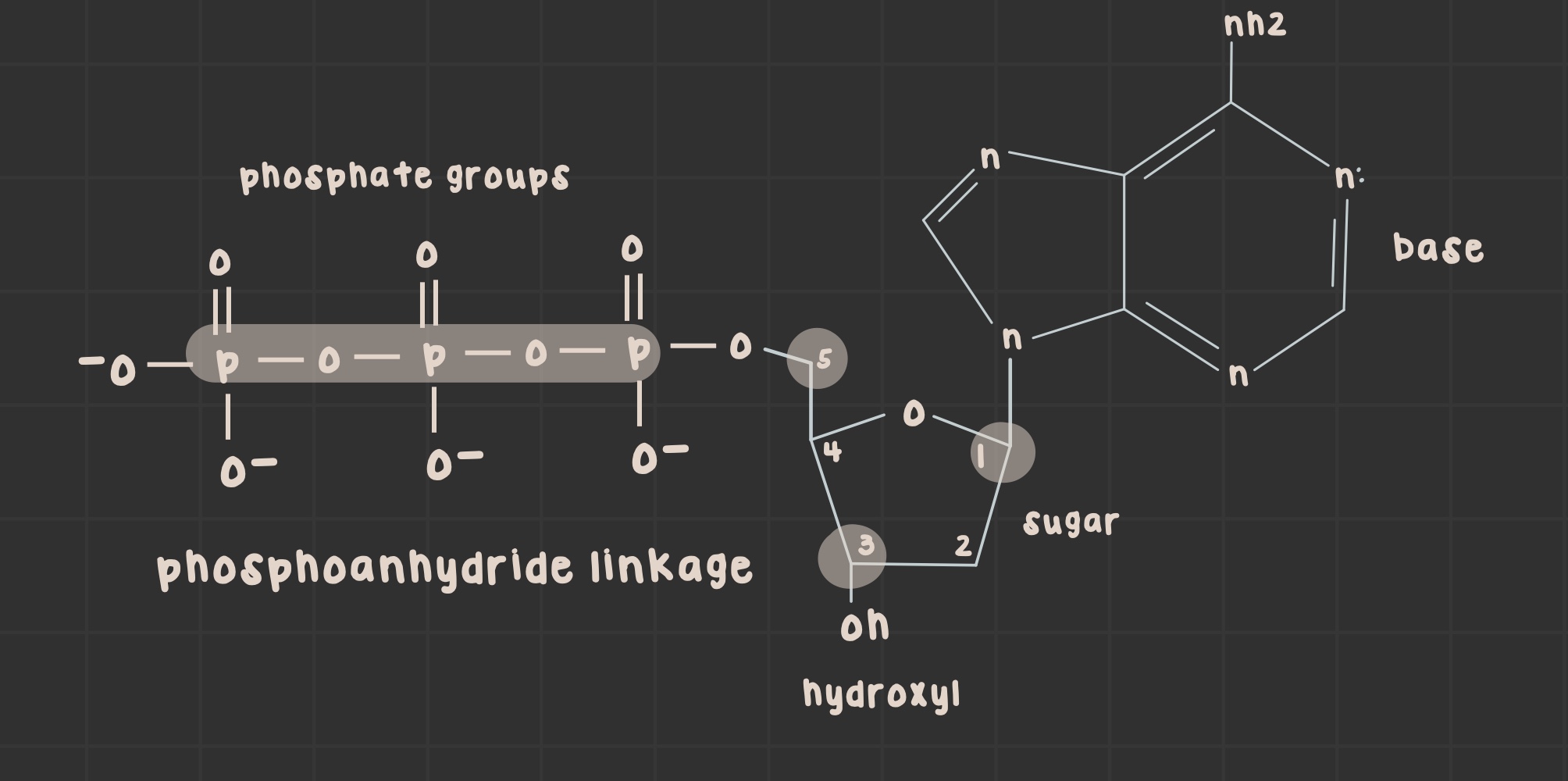
What is the sugar phosphate backbone and how is it formed?
alternating negatively charged sugar-phosphate backbone that forms as the previous 3’ OH group attacks the a-phosphate of the incoming 5’ end
What are the 5’ and 3’ ends of DNA?
5’ end: holds the free 5’ phosphate
3’ end: holds the free 3’ OH
What are the levels of organization in DNA?
primary structure: sequence of nucleotides in a nucleic acid
secondary structure: forces that hold DNA (or RNA) together; forms the double helix
hydrophobic effect occurs to protect hydrophobic bases; when exposed to water, a solvation layer forms
the double helix hides bases from water, and allows water to interact with the sugar-phosphate backbone
base pair stacking involves noncovalent interactions/pi stacking with bases to stabilize the helix
What factors impact DNA helix stability?
hydrogen bonds (more bonds = higher Tm)
longer helices (more intermolecular interactions = higher Tm)
How many hydrogen bonds are found in each nitrogenous base?
Adenine forms 2 hydrogen bonds with thymine, while cytosine forms 3 hydrogen bonds with guanine in a DNA double helix.
What factors impact DNA reannealing?
DNA length (more length = more time)
pH (closer to 7.4 = less time)
salt concentration (more cations for negative phosphates = less time)
How many origins of replication are found in eukaryotes and prokaryotes?
eukaryotes have multiple that open simultaneously (faster replication)
prokaryotes have one
What are the enzymes of DNA replication?
helicase: unwins parent helix at the origin of replication
single-stranded DNA binding proteins: hold separate strands apart for a replication bubble and two replication forks
topoisomerase: cleaves one strand to relieve strain from DNA supercoiling in front of the fork
DNA polymase: synthesizes DNA in the 5’-3’ direction while reading 3’-5’
creates the leading (3’-5’ and lagging (5’-3’) strand
primase: synthesizes a short 5'-3’ RNA primer on the leading and lagging strands
creates okazaki fragments
DNA pol III: attaches uncoupled dNTPs to the growing DNA strand after an RNA primer is placed
free dNTP enters DNA pol III catalytic site and hydrogen bonds a complementary nucleotide on the parent strand
the free 3’ OH from the previous nucleotide of the growing strand attacks the incoming dNTP 5’ phosphate
dNTP releases a pyrophosphate and initiates a condensation reaction to attach the nucleotide to the growing strand
a covalent phosphodiester bond forms
DNA pol I: removes RNA primers and replaces them with DNA fragments
DNA ligase: forms phosphodiester bonds to join all DNA fragments
Which histone proteins interact with DNA’s negative phosphates?
R and K