BCM.13 - NUCLEOTIDES
1/36
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced | Call with Kai |
|---|
No analytics yet
Send a link to your students to track their progress
37 Terms
Nucleotide
Nucleotides are molecules consisting of a nucleoside and a phosphate group.
Base + sugar + phosphate
Nucleoside
Glycosylamines that can be thought of as nucleotides without a phosphate group ( base attached to the sugar with no phosphate group )
Base + sugar
4 uses of nucleotides
1) building blocks of DNA and RNA
2) energy currency eg ATP
3) Second messengers in cell signalling (cAMP = cyclic adenosine monophosphate)
4) Electron carriers in metabolism (NAD+ = nicotinamide adenine dinucleotide)
Purines have
pyrimidines have
2 rings
1 ring
What are the two purines
Adenine and Guanine
What are the 3 pyrimidines?
cytosine, thymine, uracil
Adenine
Amino group
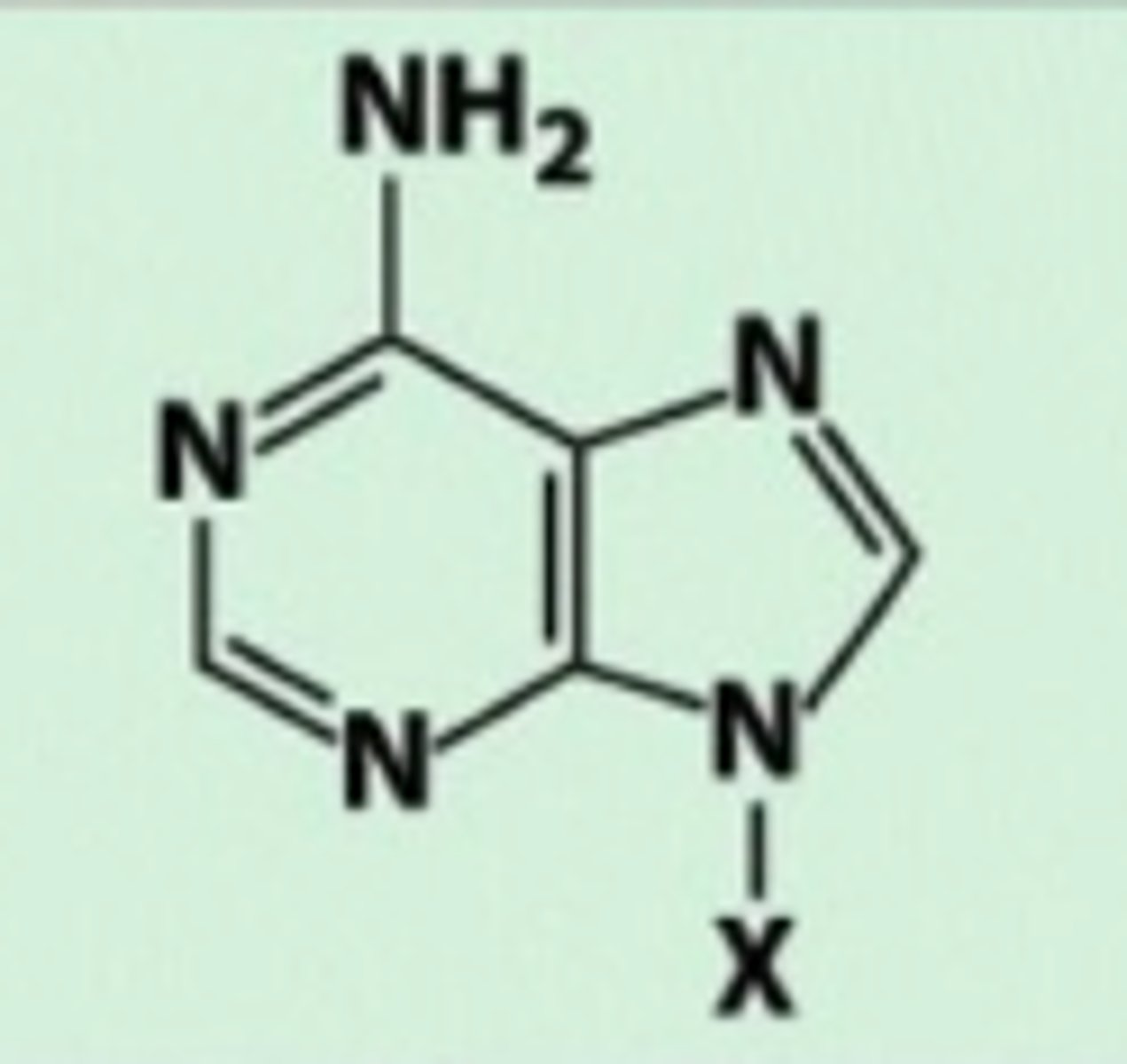
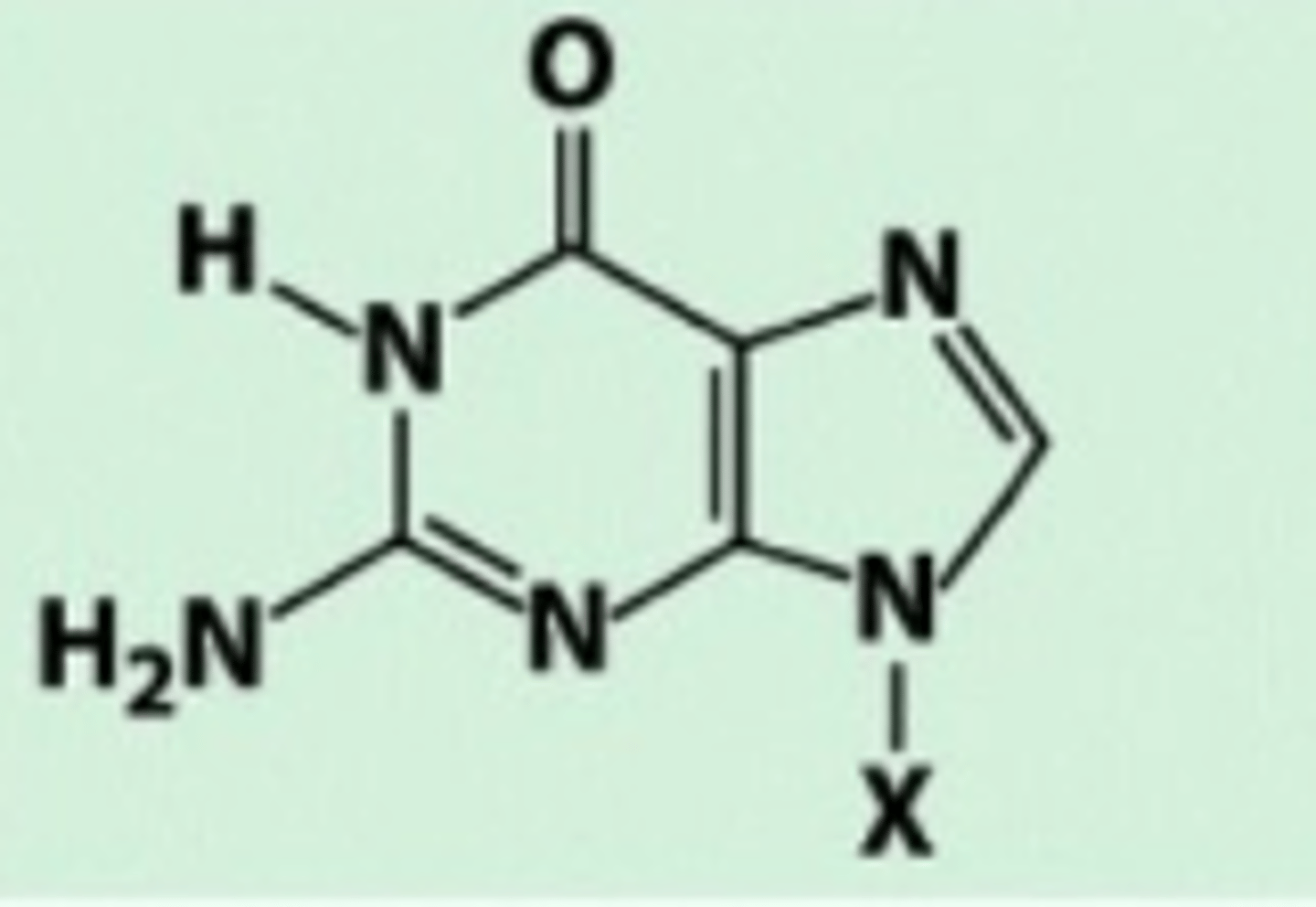
Guanine
Carbonyl group and amino group
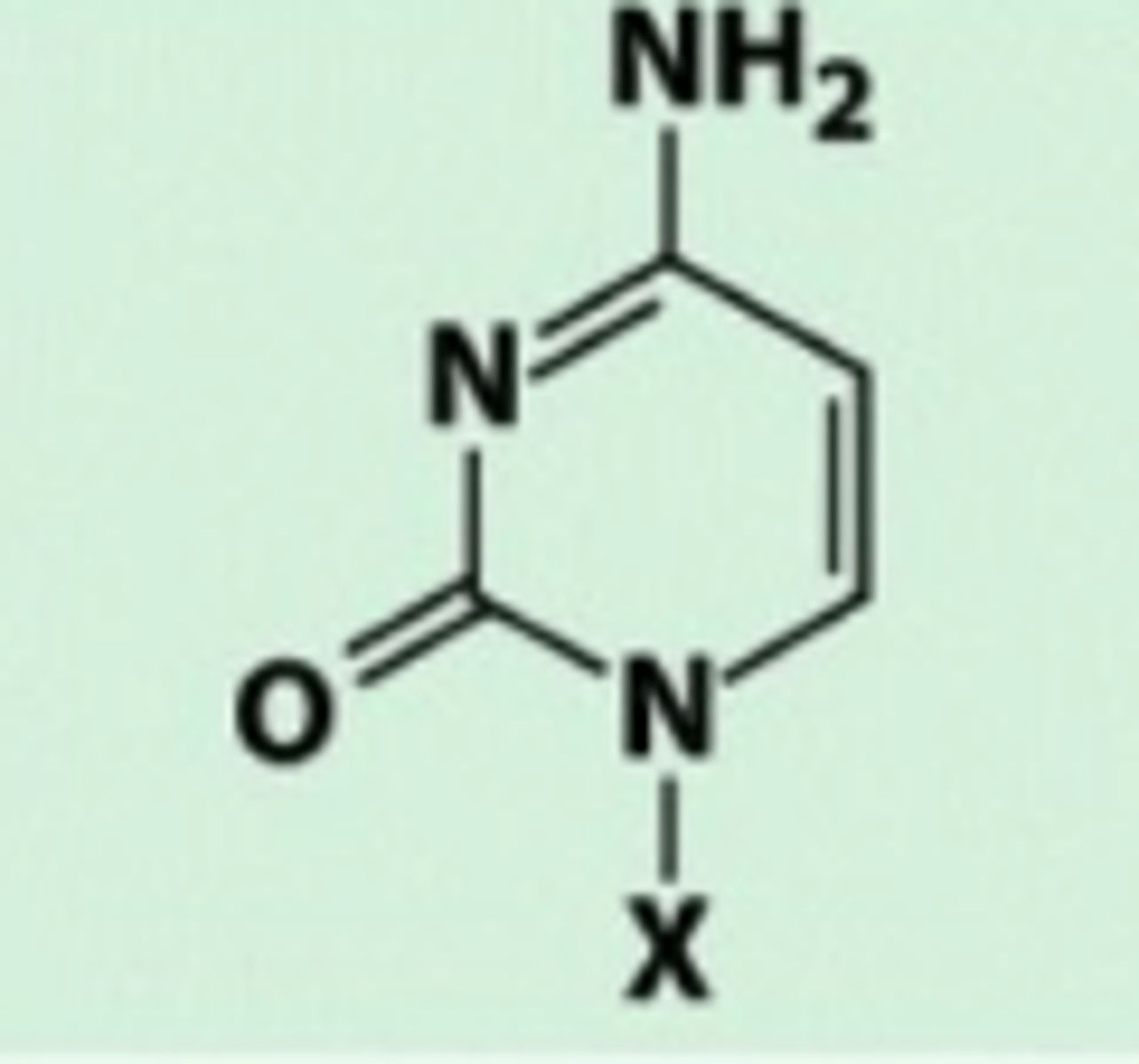
Cytosine
Amino group
Uracil
Carbonyl group
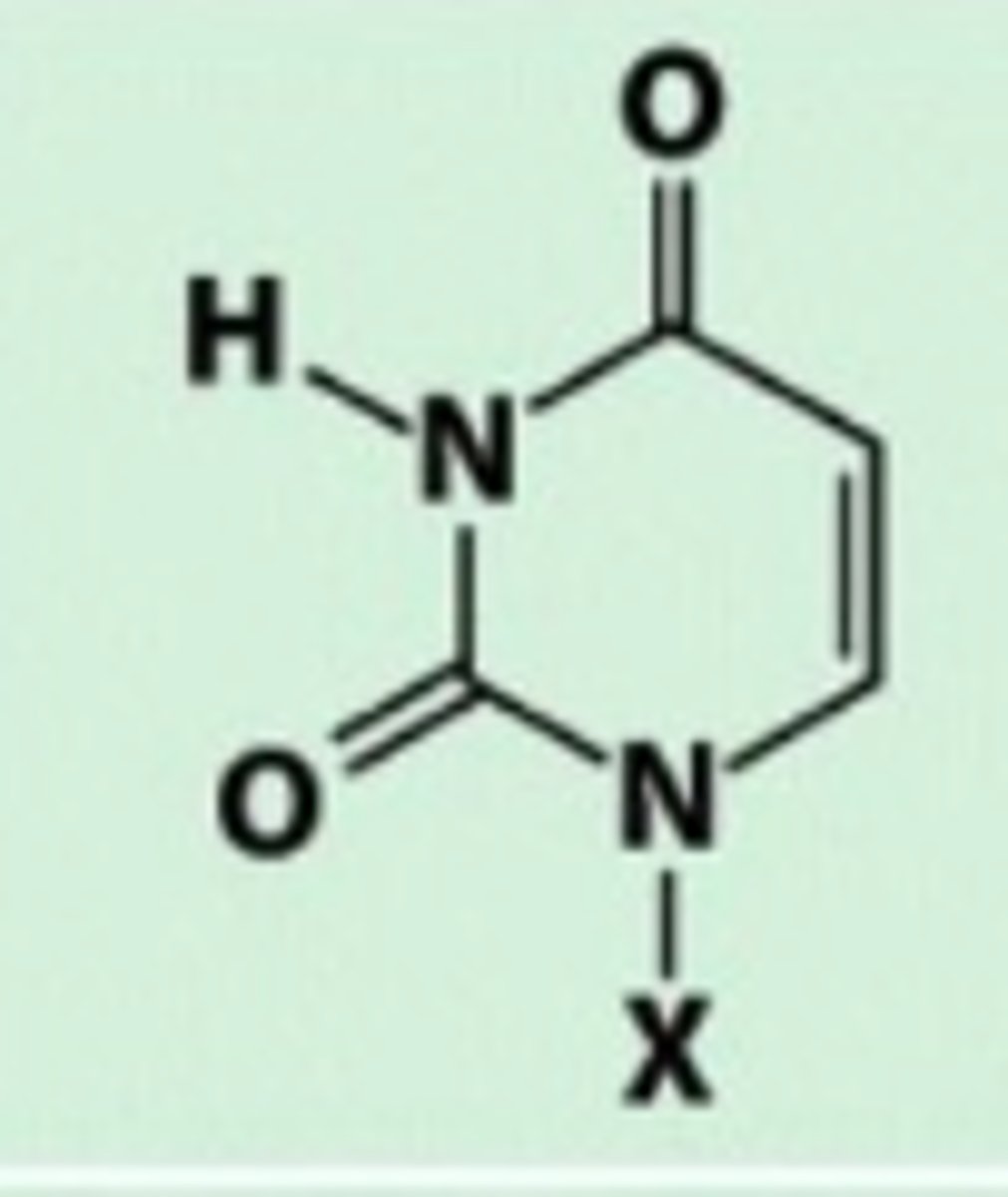
Thymine
Carbonyl group
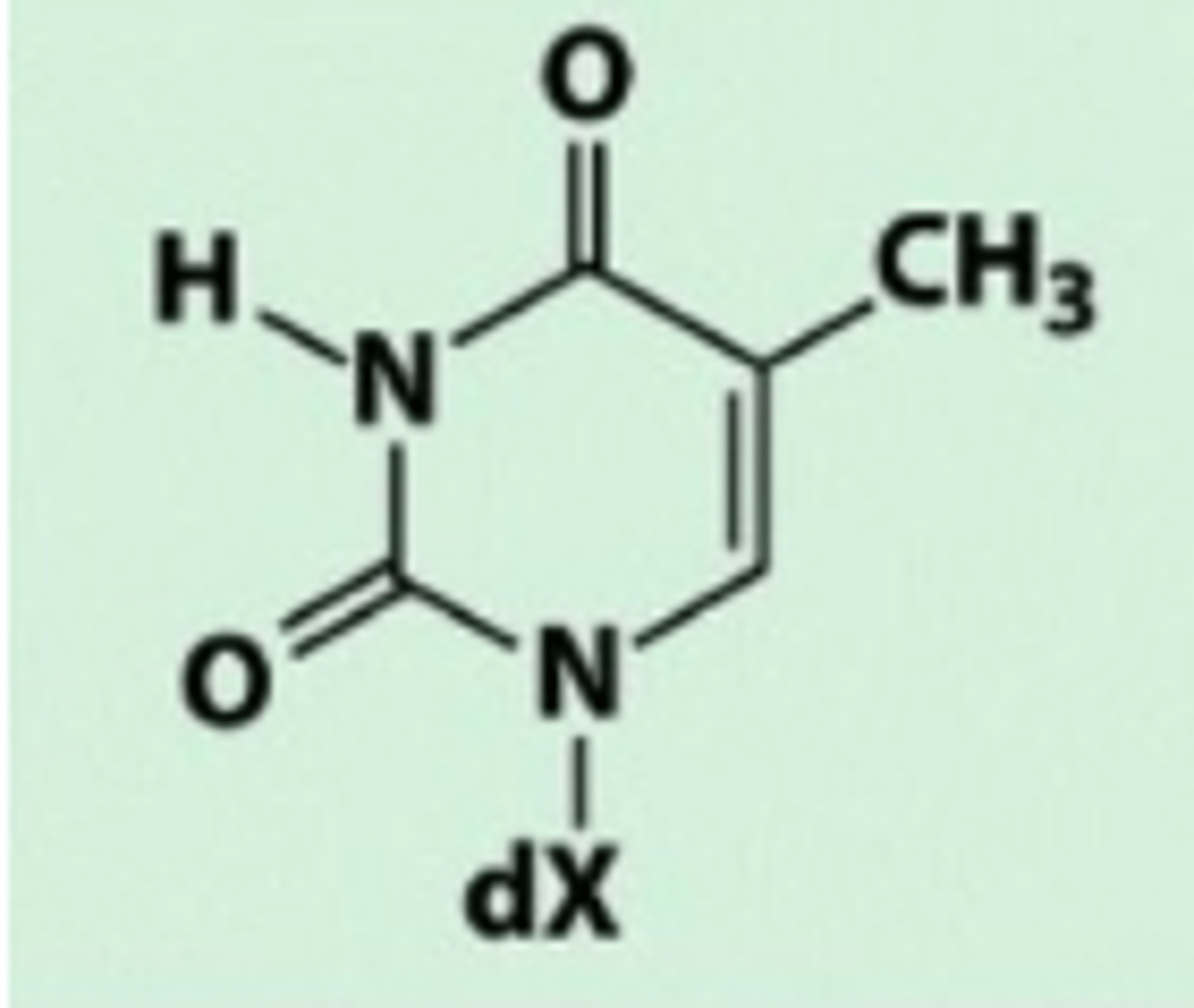
Heterocyclic
one or more non-carbon atoms in ring
Why does DNA not contain uracil
Oxidative deamination turns DNA bases into oxidised and deaminated forms.
Cell mechanisms scan the DNA for these mutated forms and cuts them out and replaces them.
However, cytosine is turned into uracil. This is a normal base so cell mechanisms cannot detect it, so if it were in DNA already uracil made from mutated cytosine would not be detected and cause severe DNA mutations that are not reversed.
Describe the covalent structure of nucleic acids.
Ribose + base joined by a beta glycosidic bond
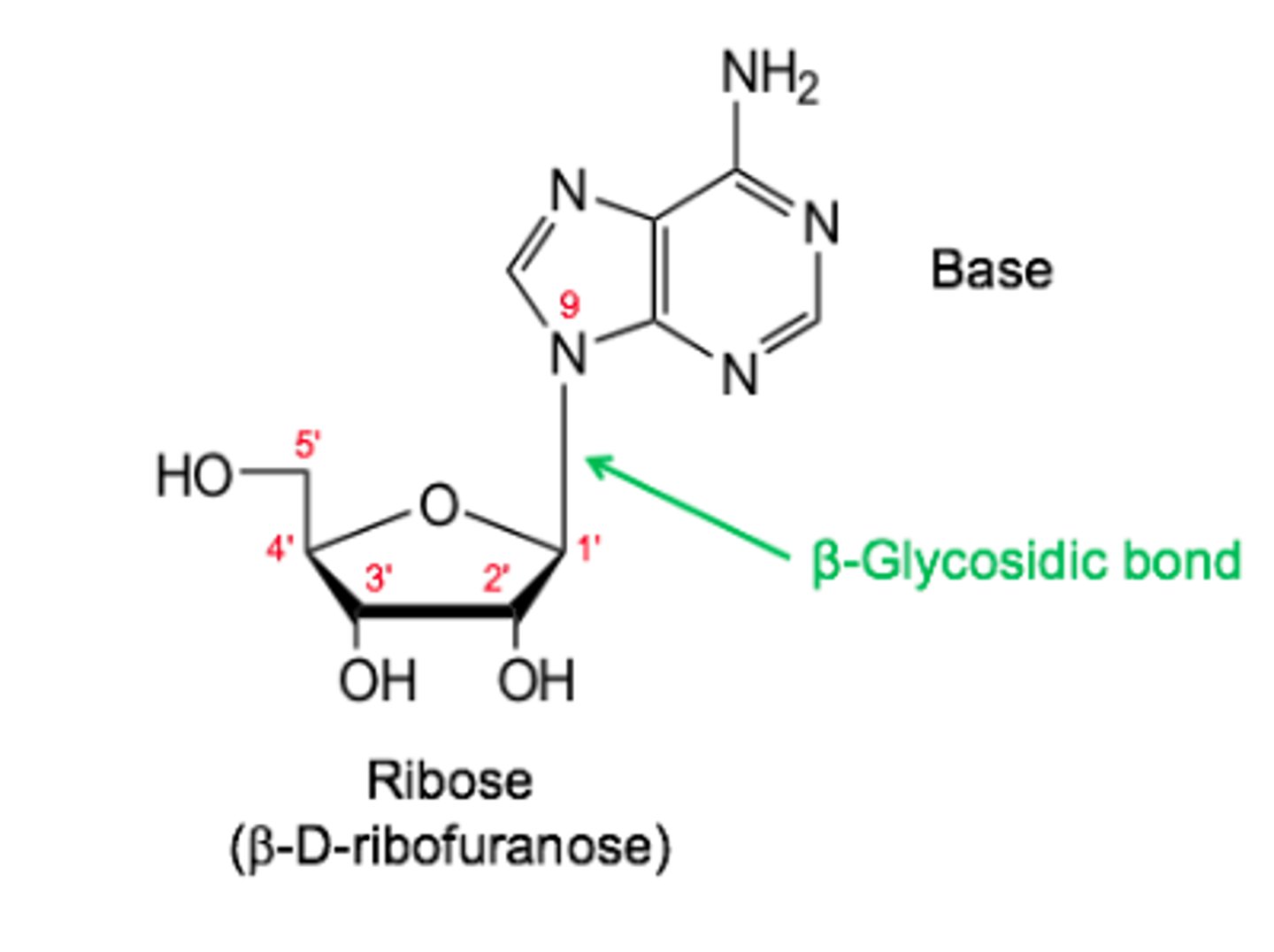
Note that primes (') are used for the numbering of the
ribose ring.
Nucleoside of adenine
Adenosine
Nucleoside of Guanine
guanosine
Nucleoside of Thymine
thymidine
Nucleoside of uracil
uridine
Nucleoside of cytosine
cytidine
DNA sequence is always written
5' to 3'.
eg d(ACG)
Uracil replaces
Thymine
Properties of RNA
1) Single stranded but often has secondary structure (hairpins, helices)
2) chemically more reactive (less stable) than DNA because of the presence of the OH groups which are very chemically reactive, hence why RNA is not found in our cells in high quantities or fossilised cells because it has degraded.
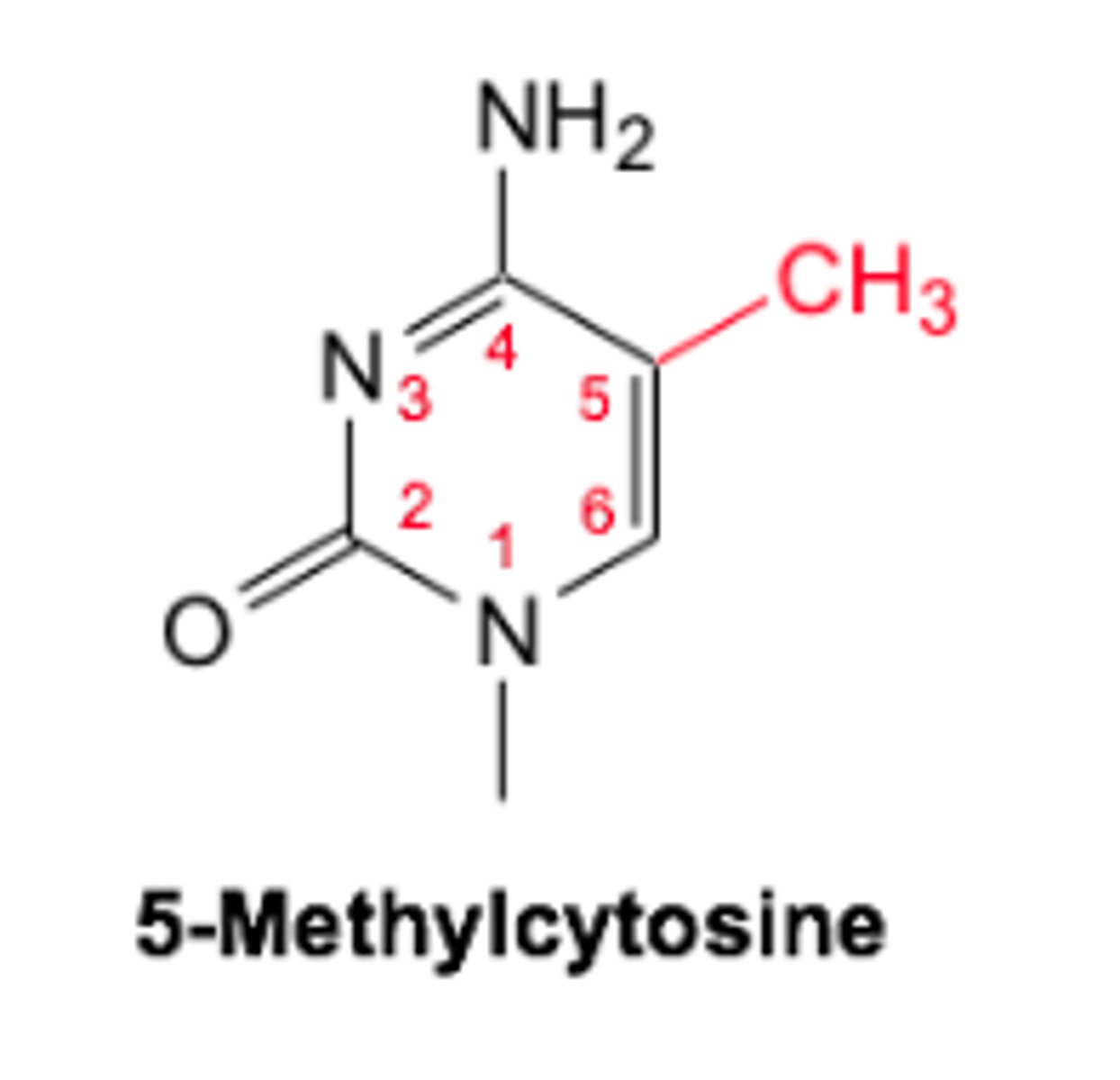
Methylated bases : 5-methylcytosine
- Occurs in mammals
- Carried out by DNA methyltransferase
- methylation of CpG sites regulates gene expression ( cytosine followed by guanine )
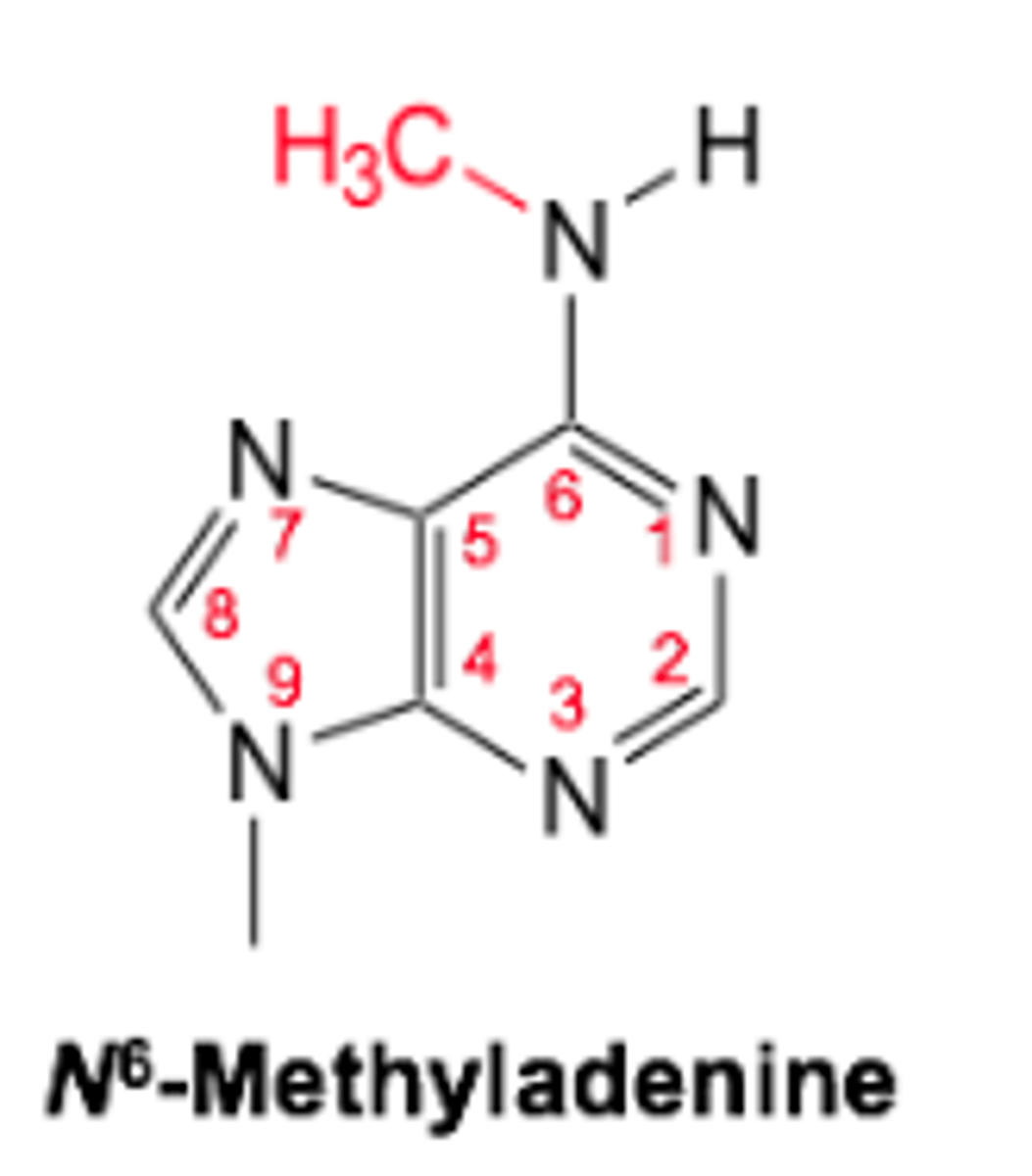
Methylated bases: N6 - methyladenine
- Occurs in bacteria
- Carried out by DNA methyltransferase
- Prevents methylation of viral DNA and ensures it is cut out of genome
- Protects DNA from digestion by restriction endonucleases
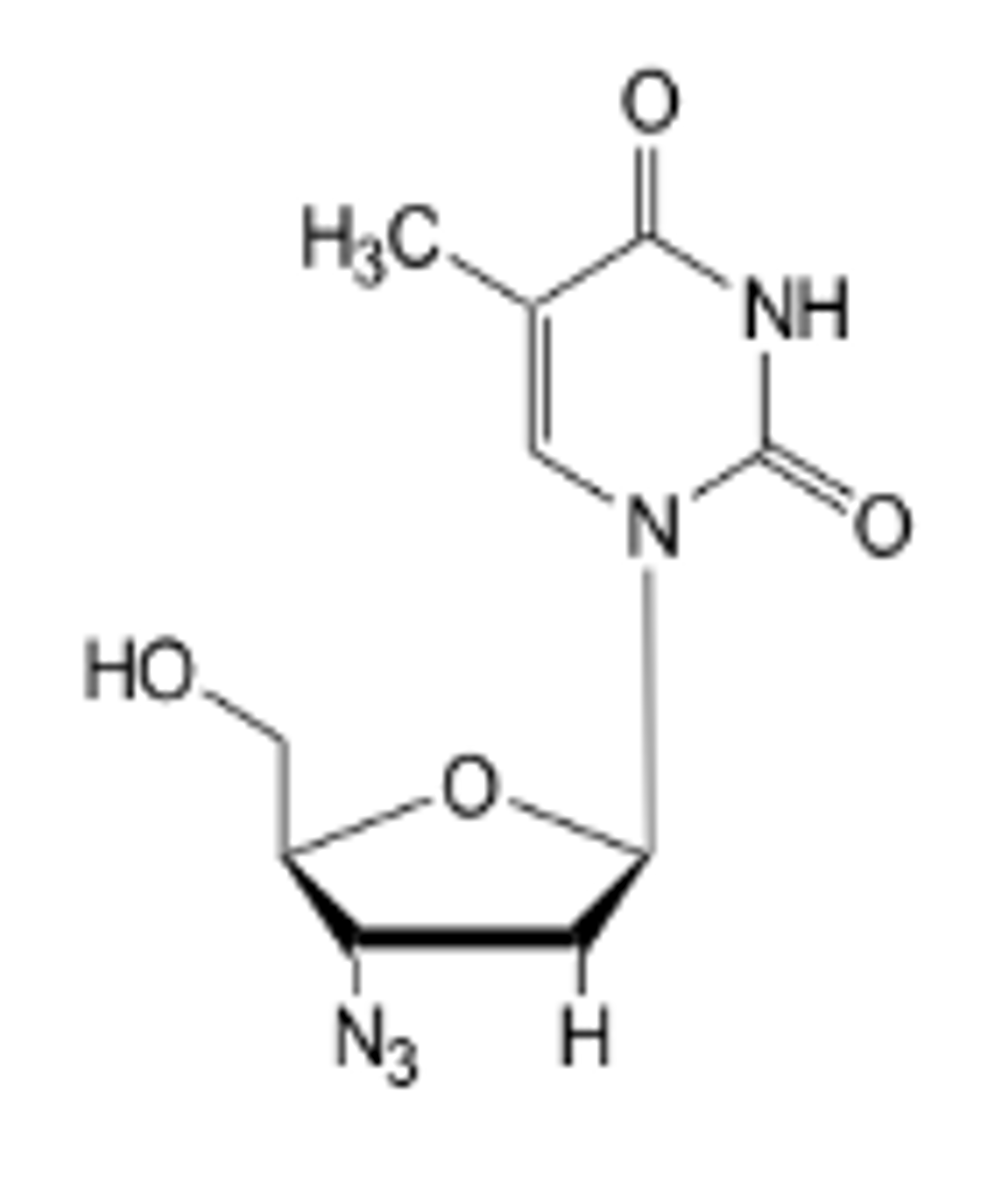
Nucleotide analogues:
Chain terminators
Man-made nucleotides that look like natural nucleotides. NO hydroxyl groups.
Binds to end of DNA chain and prevents it being extended as there are no extra OH groups to extend the chain by.
Eg AZT ( Anti HIV )
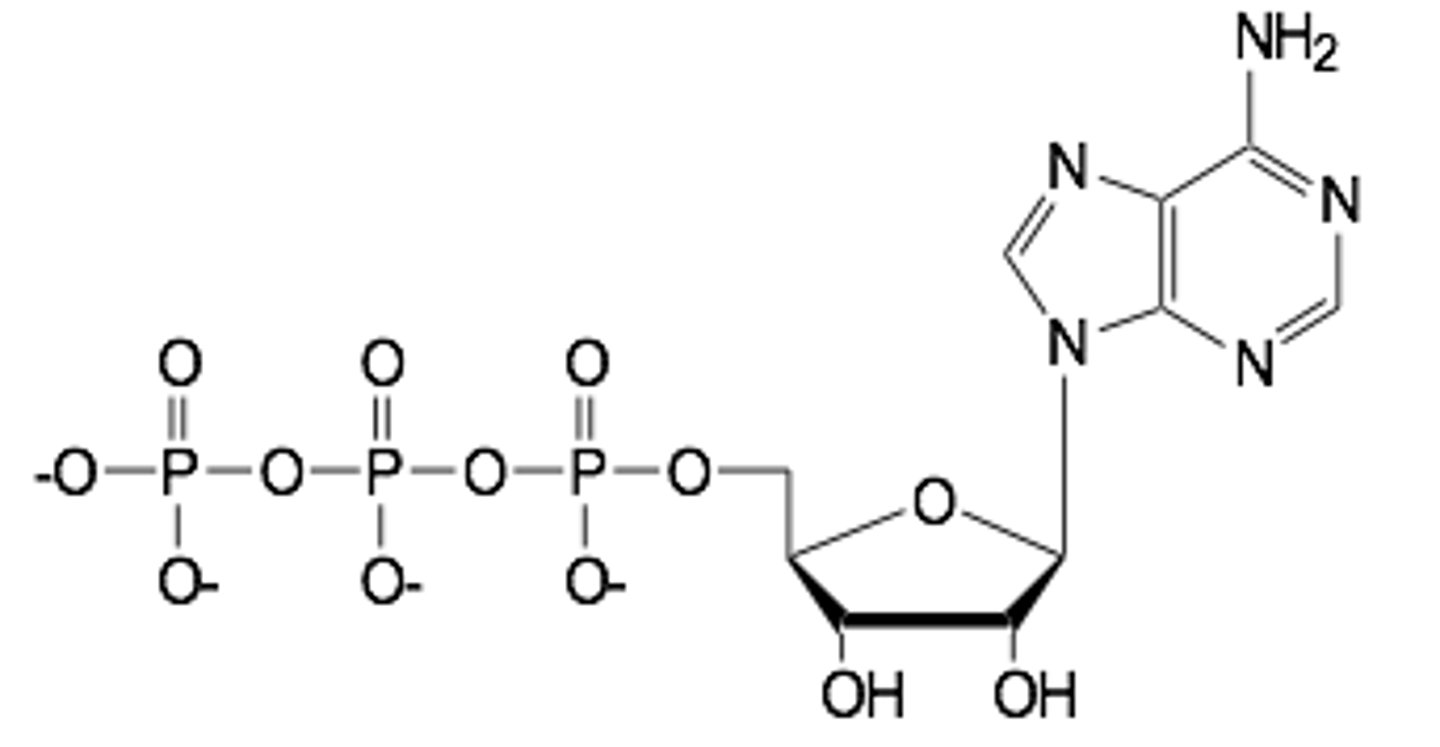
Draw ATP
5 ATP facts
1) Energy derived from the oxidation of food is converted into ATP.
2) ATP is synthesised by ATP synthase, embedded in the inner mitochondrial membrane.
3) ATP is used to drive energy-requiring processes (motion, active transport, biosynthesis).
4) Humans turn over their own body mass in ATP in one day (~100g present at any time).
5) GTP is sometimes used instead of ATP.
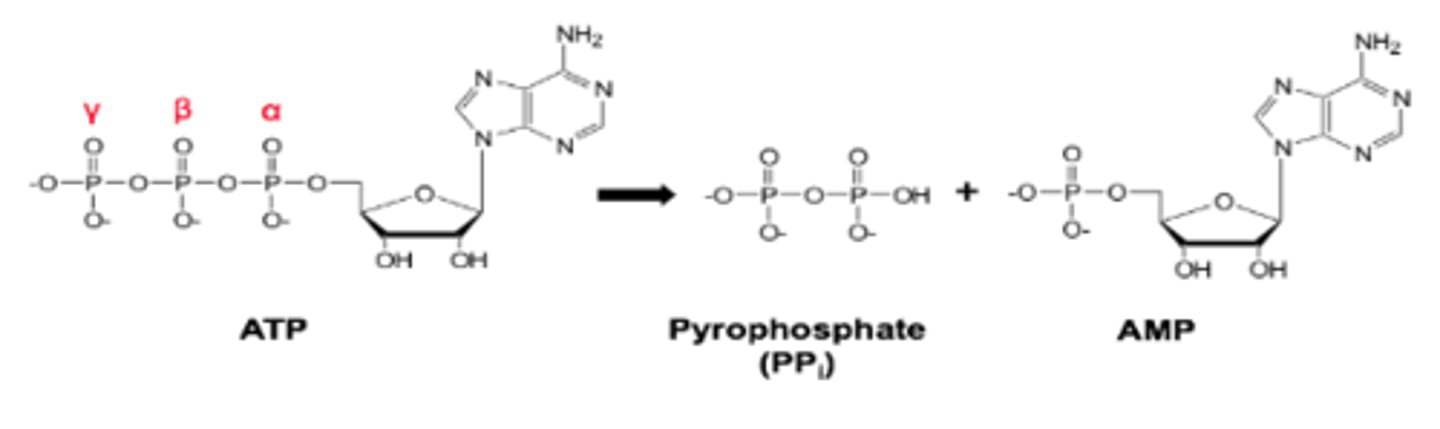
Explain how ATP hydrolysis can drive energetically unfavourable reactions.
- Coupled to energetically favourable reaction of ATP hydrolysis
-Highly exergonic process = releases a lot of energy (45.6 kJ/mol).
- Phosphoanhydride bonds (α-β and β-γ) are "high-energy" bonds.
- Hydrolysis products have less charge repulsion, higher resonance stabilisation and better solvation.
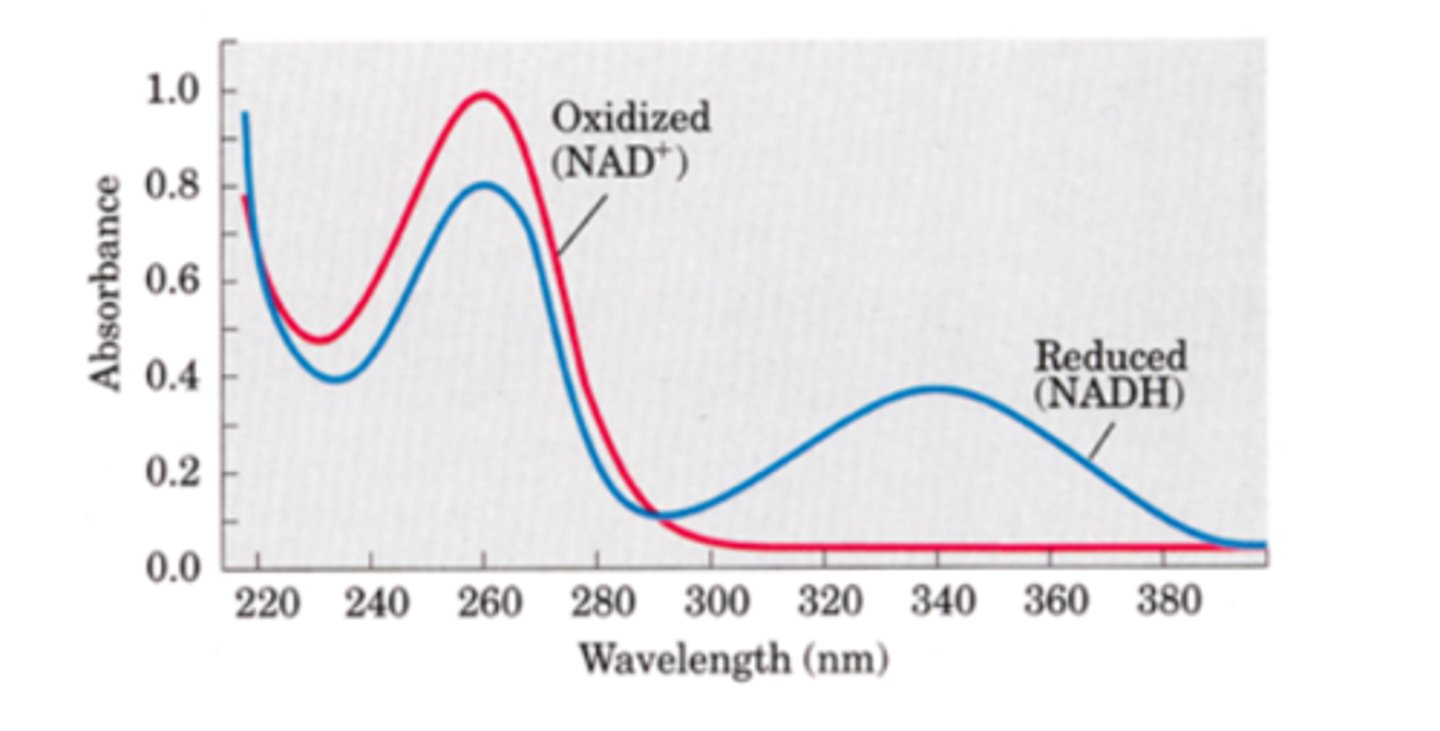
Describe the role of nucleotides that act as electron carriers
- NADH (proton added) / FADH2 ( 2 double bonds turn to one and 2 hydrogens are added )
- Switches between oxidised and reduced forms to transfer electrons between the krebs cycle and electron transport chain
NAD+ and NADH have different
absorption spectra.
Reduced NADH has a peak at the end
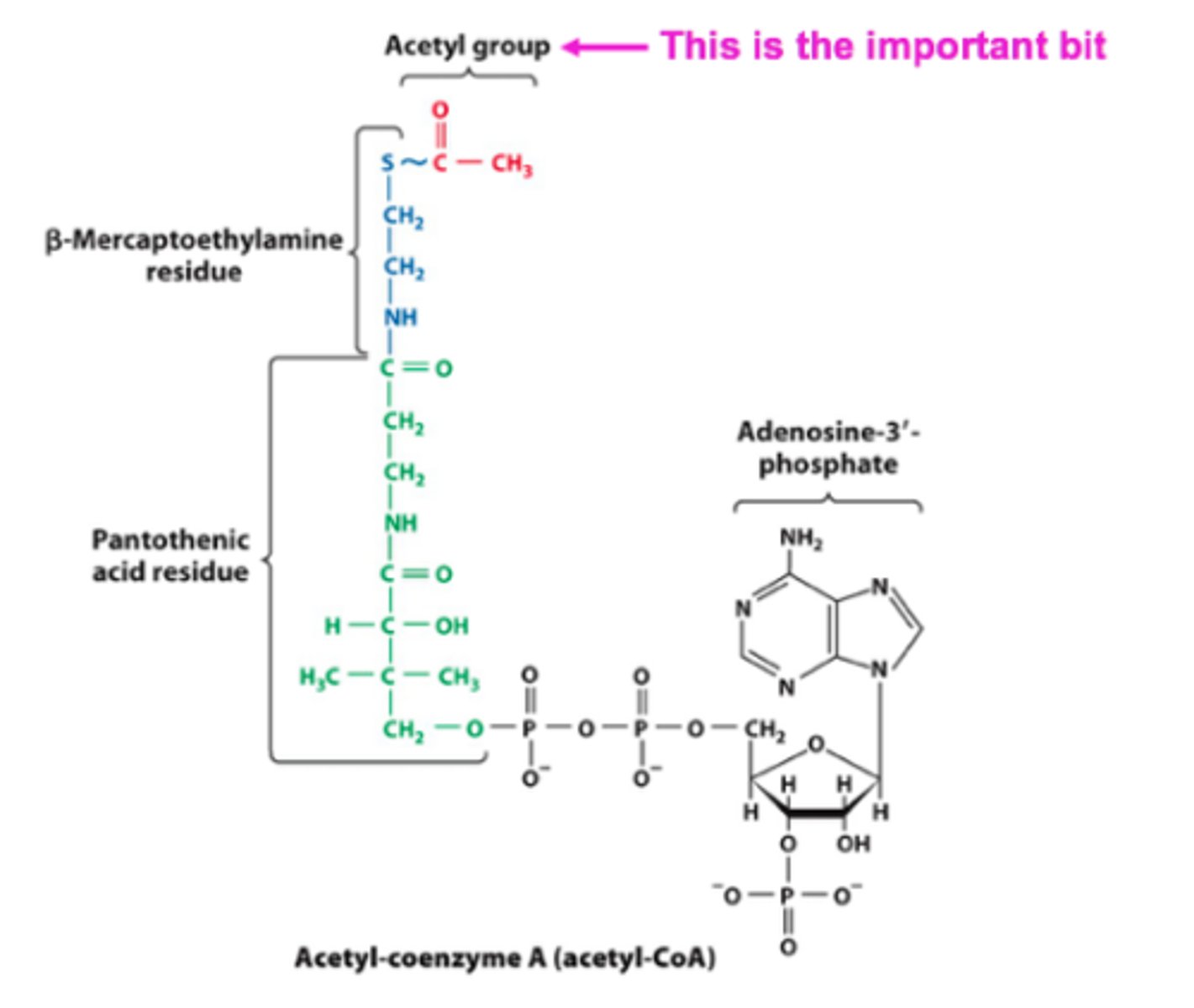
Acetyl-coenzyme A acts as a
carrier of acyl groups
Sulfur on Acetyl-coenzyme A binds to acyl groups and releases them when needed
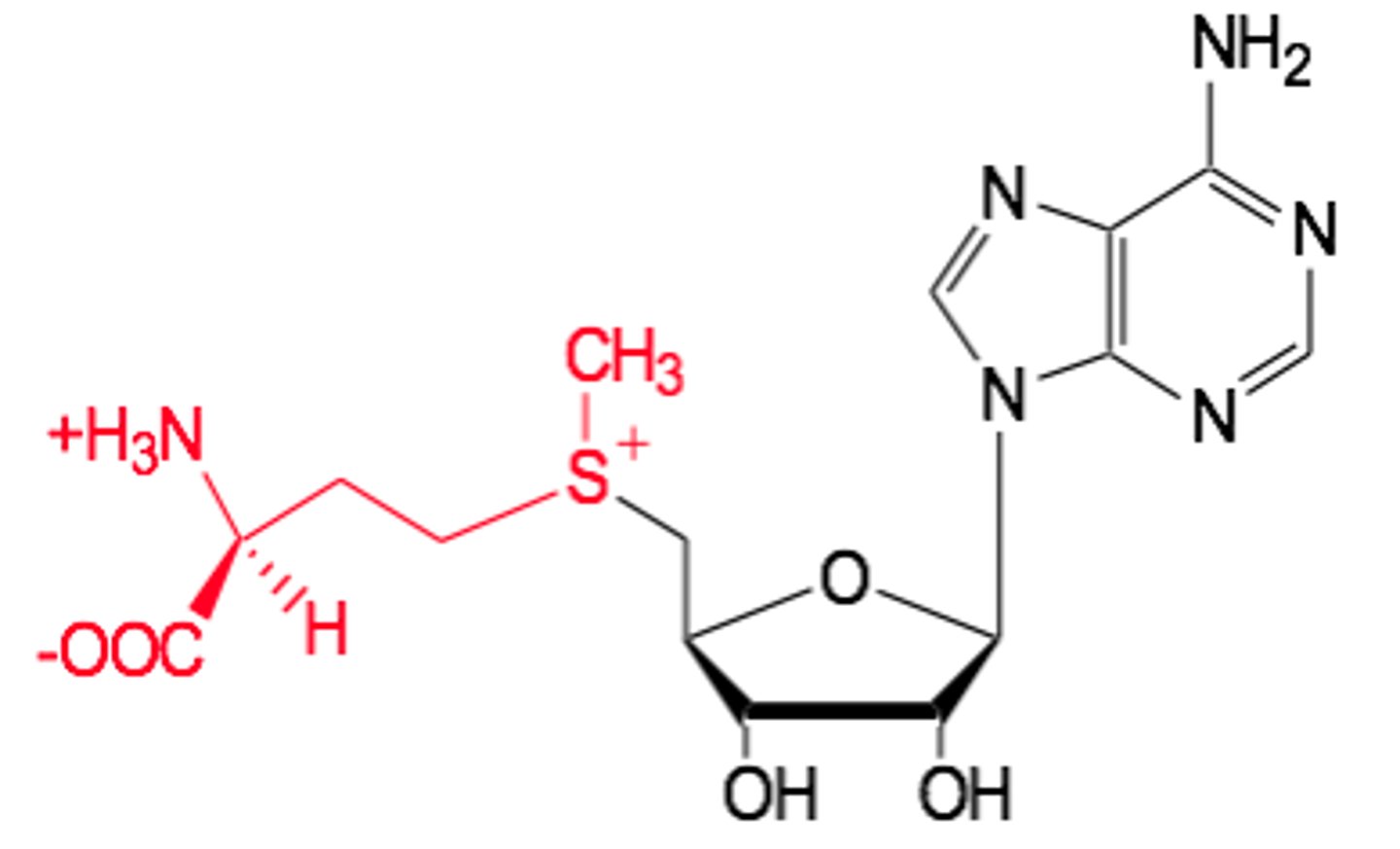
S-Adenosylmethionine acts as a
- methyl group donor
1) Synthesised from methionine and ATP by methionine adenosyl transferase (producing Pi and PPi).
2) Methyl group can be transferred to nucleic acids, proteins, lipids, and metabolites.
3) The S+-CH3 group makes the CH3 group actively want to leave the molecule attach to another compound.
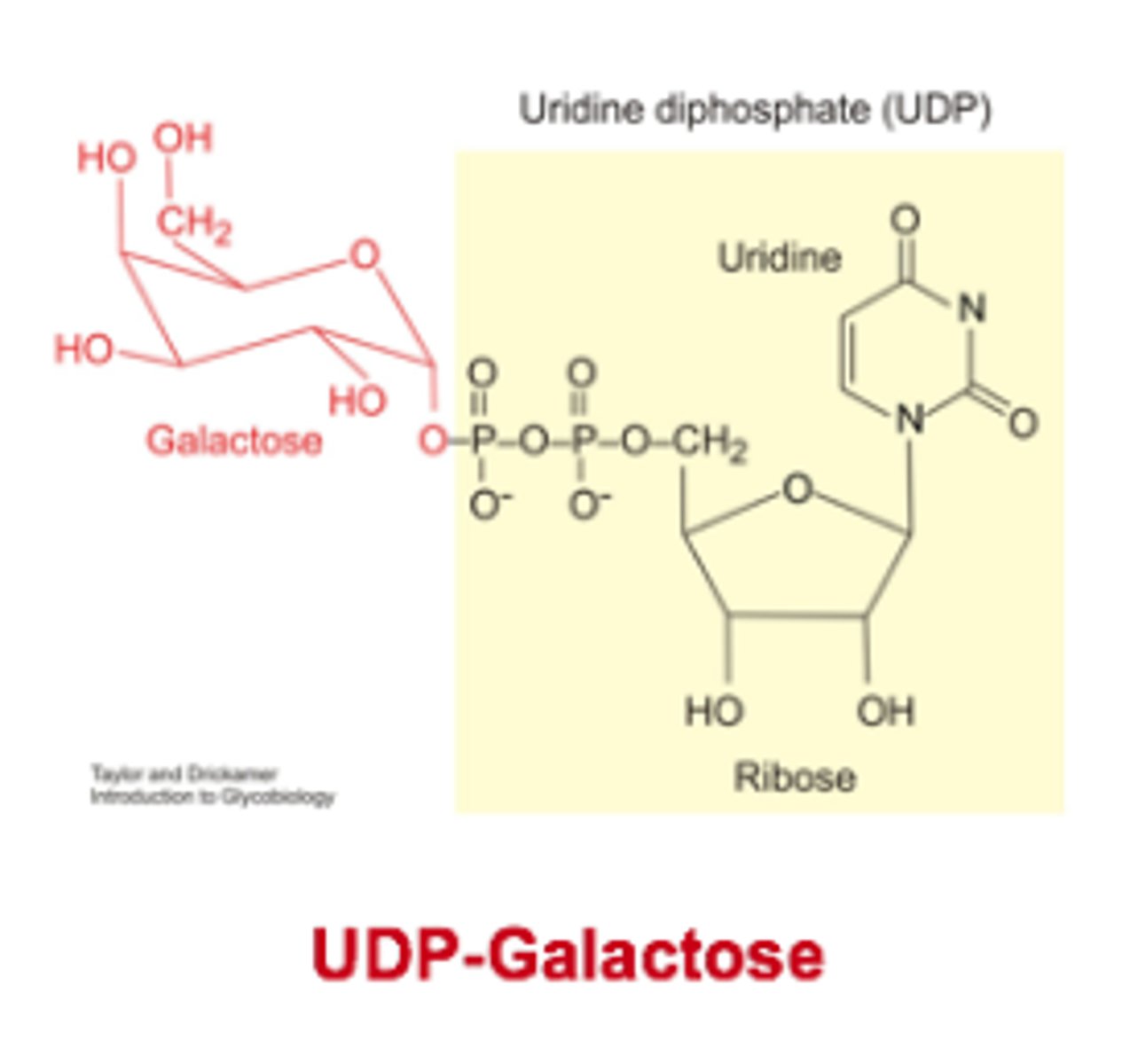
Uridine diphosphate sugars are used in
glycosidic bond formation for GLYCOGEN
1) Glycosidic bond formation requires energy. UDP-sugars are used instead of free sugars to make the reaction favourable.
2)Two ATP molecules are consumed in the synthesis of one UDP-sugar molecule meaning it contains a lot of energy.
Other functions of nucleotides:
1) Cyclic nucleotides: Signal molecules (“second messengers”) downstream of G protein-coupled receptors.
(Synthesised from corresponding NTPs by adenylyl cyclase and guanylyl cyclase, respectively, Hydrolysed by cAMP and cGMP phosphodiesterase)
2) Biosynthesis : precursors for nucleic acids and and activated intermediates eg UDP-glucose.
Examples of signalling molecules
1) Cyclic di-GMP: SM in bacteria, involved in biofilm formation
2) Cyclic AMP-GMP: SM involved in innate immunity to viruses and phages
3) Cyclic tetra-AMP: SM involved in bacterial CRISPR-Cas systems